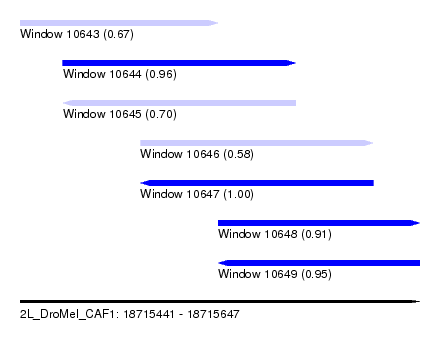
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,715,441 – 18,715,647 |
| Length | 206 |
| Max. P | 0.998912 |

| Location | 18,715,441 – 18,715,543 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 92.16 |
| Mean single sequence MFE | -29.57 |
| Consensus MFE | -29.30 |
| Energy contribution | -29.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.669662 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18715441 102 + 22407834 AUGGGGAUGGCGAUGGCGAUGGCGACGAUGGAAAACGGCGCUCCAGAUUGGCAAAAAUGUCCAGCGGAGGAGAUGUUGUUGUCGUUGACAAUGUUGCUGGUA ........(((((((.((((((((((((((.....)....((((.(.((((((....)).))))))))).....)))))))))))))....))))))).... ( -29.70) >DroSec_CAF1 8300 90 + 1 ------------AUGGCGAUGGCGACGAUGGAAAACGGCGCUCCAGAUUGGCAAAAAUGUCCAGCGGAGGAGAUGUUGUUGUCGUUGACAAUGUUGCUGGUA ------------..(((((((.(((((((....(((((((((((.(.((((((....)).)))))...)))).))))))))))))))....))))))).... ( -29.30) >DroSim_CAF1 6637 102 + 1 AUGGGGAUGGCGAUGGCGAUGGCGACGAUGGAAAACGGCGCUCCAGAUUGGCAAAAAUGUCCAGCGGAGGAGAUGUUGUUGUCGUUGACAAUGUUGCUGGUA ........(((((((.((((((((((((((.....)....((((.(.((((((....)).))))))))).....)))))))))))))....))))))).... ( -29.70) >consensus AUGGGGAUGGCGAUGGCGAUGGCGACGAUGGAAAACGGCGCUCCAGAUUGGCAAAAAUGUCCAGCGGAGGAGAUGUUGUUGUCGUUGACAAUGUUGCUGGUA ..............(((((((.(((((((....(((((((((((.(.((((((....)).)))))...)))).))))))))))))))....))))))).... (-29.30 = -29.30 + 0.00)



| Location | 18,715,463 – 18,715,583 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.50 |
| Mean single sequence MFE | -38.56 |
| Consensus MFE | -35.76 |
| Energy contribution | -35.76 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963569 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18715463 120 + 22407834 CGACGAUGGAAAACGGCGCUCCAGAUUGGCAAAAAUGUCCAGCGGAGGAGAUGUUGUUGUCGUUGACAAUGUUGCUGGUACGGGAUUGCAGUUGCAACUGCAACUGCACUGAGGAAAUAG (((((((....(((((((((((.(.((((((....)).)))))...)))).))))))))))))))....((((.((....(((...(((((((((....)))))))))))).)).)))). ( -38.80) >DroSec_CAF1 8310 120 + 1 CGACGAUGGAAAACGGCGCUCCAGAUUGGCAAAAAUGUCCAGCGGAGGAGAUGUUGUUGUCGUUGACAAUGUUGCUGGUACGGGAUUGCAGUUGCAACUGCAACUGCACUGAGGAAAUAG (((((((....(((((((((((.(.((((((....)).)))))...)))).))))))))))))))....((((.((....(((...(((((((((....)))))))))))).)).)))). ( -38.80) >DroSim_CAF1 6659 120 + 1 CGACGAUGGAAAACGGCGCUCCAGAUUGGCAAAAAUGUCCAGCGGAGGAGAUGUUGUUGUCGUUGACAAUGUUGCUGGUACGGGAUUGCAGUUGCAACUGCAACUGCACUGAGGAAAUAG (((((((....(((((((((((.(.((((((....)).)))))...)))).))))))))))))))....((((.((....(((...(((((((((....)))))))))))).)).)))). ( -38.80) >DroEre_CAF1 8292 120 + 1 CGACGAUGGAAAACUGCGCUCCAGAUUGGCAAAAAUGUCCAGCGGAGGAGAUGUUGUUGCCGUUGACAAUGUUGCUGCUACGGGAUUGCAGUUGCAACUGCAACUGCACUGAGGAAACAG ...((((((..(((.(((((((.(.((((((....)).)))))...)))).))).))).))))))....((((....((.(((...(((((((((....)))))))))))).)).)))). ( -39.10) >DroYak_CAF1 8310 120 + 1 CGACGAUGGAAAACGGCGCUCCAAAUUGGCAAAAAUGUACAGCGGAGGAGAUGUUGUUGCCGUUGACAAUGUUGCUGGAACGGGAUUGCAGUUGCAACUGCAACUGCACUGAGGAAAUAG ((.((.(....).)).)).(((...(..((((...(((.((((((..(........)..)))))))))...))))..)..(((...(((((((((....)))))))))))).)))..... ( -37.30) >consensus CGACGAUGGAAAACGGCGCUCCAGAUUGGCAAAAAUGUCCAGCGGAGGAGAUGUUGUUGUCGUUGACAAUGUUGCUGGUACGGGAUUGCAGUUGCAACUGCAACUGCACUGAGGAAAUAG ...((((((..(((((((((((...((((((....)).))))....)))).))))))).))))))....((((.((....(((...(((((((((....)))))))))))).)).)))). (-35.76 = -35.76 + 0.00)



| Location | 18,715,463 – 18,715,583 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.50 |
| Mean single sequence MFE | -29.72 |
| Consensus MFE | -26.04 |
| Energy contribution | -26.00 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.704304 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18715463 120 - 22407834 CUAUUUCCUCAGUGCAGUUGCAGUUGCAACUGCAAUCCCGUACCAGCAACAUUGUCAACGACAACAACAUCUCCUCCGCUGGACAUUUUUGCCAAUCUGGAGCGCCGUUUUCCAUCGUCG ............(((((((((....)))))))))....(((.((((.....(((((...)))))...............(((.((....)))))..)))).)))................ ( -28.10) >DroSec_CAF1 8310 120 - 1 CUAUUUCCUCAGUGCAGUUGCAGUUGCAACUGCAAUCCCGUACCAGCAACAUUGUCAACGACAACAACAUCUCCUCCGCUGGACAUUUUUGCCAAUCUGGAGCGCCGUUUUCCAUCGUCG ............(((((((((....)))))))))....(((.((((.....(((((...)))))...............(((.((....)))))..)))).)))................ ( -28.10) >DroSim_CAF1 6659 120 - 1 CUAUUUCCUCAGUGCAGUUGCAGUUGCAACUGCAAUCCCGUACCAGCAACAUUGUCAACGACAACAACAUCUCCUCCGCUGGACAUUUUUGCCAAUCUGGAGCGCCGUUUUCCAUCGUCG ............(((((((((....)))))))))....(((.((((.....(((((...)))))...............(((.((....)))))..)))).)))................ ( -28.10) >DroEre_CAF1 8292 120 - 1 CUGUUUCCUCAGUGCAGUUGCAGUUGCAACUGCAAUCCCGUAGCAGCAACAUUGUCAACGGCAACAACAUCUCCUCCGCUGGACAUUUUUGCCAAUCUGGAGCGCAGUUUUCCAUCGUCG ((((((((....(((((((((....)))))))))...........((((...((((..((((...............))))))))...))))......)))).))))............. ( -31.96) >DroYak_CAF1 8310 120 - 1 CUAUUUCCUCAGUGCAGUUGCAGUUGCAACUGCAAUCCCGUUCCAGCAACAUUGUCAACGGCAACAACAUCUCCUCCGCUGUACAUUUUUGCCAAUUUGGAGCGCCGUUUUCCAUCGUCG ............(((((((((....)))))))))....(((((((((((...(((..(((((...............))))))))...)))).....)))))))................ ( -32.36) >consensus CUAUUUCCUCAGUGCAGUUGCAGUUGCAACUGCAAUCCCGUACCAGCAACAUUGUCAACGACAACAACAUCUCCUCCGCUGGACAUUUUUGCCAAUCUGGAGCGCCGUUUUCCAUCGUCG ............(((((((((....)))))))))...........((....(((((...))))).........(((((.(((.((....)))))...))))).))............... (-26.04 = -26.00 + -0.04)


| Location | 18,715,503 – 18,715,623 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.00 |
| Mean single sequence MFE | -38.40 |
| Consensus MFE | -34.06 |
| Energy contribution | -34.14 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.584854 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18715503 120 + 22407834 AGCGGAGGAGAUGUUGUUGUCGUUGACAAUGUUGCUGGUACGGGAUUGCAGUUGCAACUGCAACUGCACUGAGGAAAUAGCCCAGCGCCACCAUUACUGUGGCCUGAAAUGUUGAAGUGU ..(((....(((((((((......)))))))))((((...(((...(((((((((....)))))))))))).((......))))))(((((.......)))))))).............. ( -36.70) >DroSec_CAF1 8350 120 + 1 AGCGGAGGAGAUGUUGUUGUCGUUGACAAUGUUGCUGGUACGGGAUUGCAGUUGCAACUGCAACUGCACUGAGGAAAUAGCCCAGCGCCACCAUUACUGUGGUCAAAAAUGUUAAAGUGU ((((..((.(((((((((......)))))))))((((...(((...(((((((((....)))))))))))).((......)))))).))(((((....)))))......))))....... ( -35.20) >DroSim_CAF1 6699 120 + 1 AGCGGAGGAGAUGUUGUUGUCGUUGACAAUGUUGCUGGUACGGGAUUGCAGUUGCAACUGCAACUGCACUGAGGAAAUAGCCCAGCGCCACCAUUACUGUGGCCAAAAAUAUUGAAGUGU .((.((.((.(......).)).))..(((((((((((...(((...(((((((((....)))))))))))).((......))))))(((((.......)))))....)))))))..)).. ( -36.00) >DroEre_CAF1 8332 120 + 1 AGCGGAGGAGAUGUUGUUGCCGUUGACAAUGUUGCUGCUACGGGAUUGCAGUUGCAACUGCAACUGCACUGAGGAAACAGCCCAGCGCCAACAUUCCUGUGGCCUAAAAUGUUGAACUGU .((((.((..((((((((......))))))))..))(((((((((.(((((((((....)))))))))(((.((......))))).........))))))))).............)))) ( -42.90) >DroYak_CAF1 8350 120 + 1 AGCGGAGGAGAUGUUGUUGCCGUUGACAAUGUUGCUGGAACGGGAUUGCAGUUGCAACUGCAACUGCACUGAGGAAAUAGCCCAGCGCCAACAUUCCUGUGGCUAAAAACGUUGCAGUGC .((......(((((((((......)))))))))((((.((((....(((((((((....)))))))))(((.((......))))).((((.((....))))))......)))).)))))) ( -41.20) >consensus AGCGGAGGAGAUGUUGUUGUCGUUGACAAUGUUGCUGGUACGGGAUUGCAGUUGCAACUGCAACUGCACUGAGGAAAUAGCCCAGCGCCACCAUUACUGUGGCCAAAAAUGUUGAAGUGU .((...((..((((((((......))))))))..)).....(((..(((((((((....)))))))))(((......)))))).))(((((.......)))))................. (-34.06 = -34.14 + 0.08)


| Location | 18,715,503 – 18,715,623 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.00 |
| Mean single sequence MFE | -36.41 |
| Consensus MFE | -31.20 |
| Energy contribution | -31.36 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.86 |
| SVM decision value | 3.28 |
| SVM RNA-class probability | 0.998912 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18715503 120 - 22407834 ACACUUCAACAUUUCAGGCCACAGUAAUGGUGGCGCUGGGCUAUUUCCUCAGUGCAGUUGCAGUUGCAACUGCAAUCCCGUACCAGCAACAUUGUCAACGACAACAACAUCUCCUCCGCU .................(((((.......)))))((((((((........)))((((((((....)))))))).........)))))....(((((...)))))................ ( -34.50) >DroSec_CAF1 8350 120 - 1 ACACUUUAACAUUUUUGACCACAGUAAUGGUGGCGCUGGGCUAUUUCCUCAGUGCAGUUGCAGUUGCAACUGCAAUCCCGUACCAGCAACAUUGUCAACGACAACAACAUCUCCUCCGCU .........((((.(((....))).))))((((.((((((((........)))((((((((....)))))))).........)))))....(((((...)))))...........)))). ( -33.00) >DroSim_CAF1 6699 120 - 1 ACACUUCAAUAUUUUUGGCCACAGUAAUGGUGGCGCUGGGCUAUUUCCUCAGUGCAGUUGCAGUUGCAACUGCAAUCCCGUACCAGCAACAUUGUCAACGACAACAACAUCUCCUCCGCU .................(((((.......)))))((((((((........)))((((((((....)))))))).........)))))....(((((...)))))................ ( -34.50) >DroEre_CAF1 8332 120 - 1 ACAGUUCAACAUUUUAGGCCACAGGAAUGUUGGCGCUGGGCUGUUUCCUCAGUGCAGUUGCAGUUGCAACUGCAAUCCCGUAGCAGCAACAUUGUCAACGGCAACAACAUCUCCUCCGCU ......((((((((..(....)..))))))))(((..((((((((.......(((((((((....))))))))).......))))))....((((.....))))........))..))). ( -41.74) >DroYak_CAF1 8350 120 - 1 GCACUGCAACGUUUUUAGCCACAGGAAUGUUGGCGCUGGGCUAUUUCCUCAGUGCAGUUGCAGUUGCAACUGCAAUCCCGUUCCAGCAACAUUGUCAACGGCAACAACAUCUCCUCCGCU ................(((...((((((((((..((((((((........)))((((((((....)))))))).........)))))....((((.....)))))))))).))))..))) ( -38.30) >consensus ACACUUCAACAUUUUUGGCCACAGUAAUGGUGGCGCUGGGCUAUUUCCUCAGUGCAGUUGCAGUUGCAACUGCAAUCCCGUACCAGCAACAUUGUCAACGACAACAACAUCUCCUCCGCU .................((((((....)).))))((((((((........)))((((((((....)))))))).........)))))....(((((...)))))................ (-31.20 = -31.36 + 0.16)


| Location | 18,715,543 – 18,715,647 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.17 |
| Mean single sequence MFE | -33.38 |
| Consensus MFE | -25.82 |
| Energy contribution | -26.06 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.907448 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18715543 104 + 22407834 CGGGAUUGCAGUUGCAACUGCAACUGCACUGAGGAAAUAGCCCAGCGCCACCAUUACUGUGGCCUGAAAUGUUGAAGUGUUAUUUGUAACCACCAUGUUCUCCA---------------- ..(((.(((((((((....)))))))))....((.((((...(((.(((((.......))))))))...))))...(.((((....))))).))......))).---------------- ( -32.50) >DroSec_CAF1 8390 104 + 1 CGGGAUUGCAGUUGCAACUGCAACUGCACUGAGGAAAUAGCCCAGCGCCACCAUUACUGUGGUCAAAAAUGUUAAAGUGUUAUUUGUAACCACCUUGUUCUUAU---------------- .((...(((((((((....)))))))))(((.((......))))).....))......(((((...(((((.........)))))...)))))...........---------------- ( -27.90) >DroSim_CAF1 6739 117 + 1 CGGGAUUGCAGUUGCAACUGCAACUGCACUGAGGAAAUAGCCCAGCGCCACCAUUACUGUGGCCAAAAAUAUUGAAGUGUUAUUUGCAACCACCAUGUUCUCAUUUAAAACCGAUUU--- (((...(((((((((....))))))))).((((((.........(((((((.......)))))....(((((....)))))....))..........)))))).......)))....--- ( -32.71) >DroEre_CAF1 8372 120 + 1 CGGGAUUGCAGUUGCAACUGCAACUGCACUGAGGAAACAGCCCAGCGCCAACAUUCCUGUGGCCUAAAAUGUUGAACUGUGAUUUGCAACCACCUGGUUCUUAUUUUAAAUCACUUUGGA .(((..(((((((((....)))))))))....(....)..)))...........(((.((((..(((((((..((((((((.........)))..))))).)))))))..))))...))) ( -37.00) >DroYak_CAF1 8390 120 + 1 CGGGAUUGCAGUUGCAACUGCAACUGCACUGAGGAAAUAGCCCAGCGCCAACAUUCCUGUGGCUAAAAACGUUGCAGUGCGAGUUGCAACCAUCUUGUUCUUAUGUUGAAUCACUUUGGA ((((((....((((((((((((.((((((((.((......))))).((((.((....)))))).........)))))))).))))))))).))))))....................... ( -36.80) >consensus CGGGAUUGCAGUUGCAACUGCAACUGCACUGAGGAAAUAGCCCAGCGCCACCAUUACUGUGGCCAAAAAUGUUGAAGUGUUAUUUGCAACCACCUUGUUCUUAU_U__AA_C__UUU___ .((...(((((((((....)))))))))(((.((......))))).(((((.......)))))..........................))............................. (-25.82 = -26.06 + 0.24)



| Location | 18,715,543 – 18,715,647 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.17 |
| Mean single sequence MFE | -35.34 |
| Consensus MFE | -28.22 |
| Energy contribution | -28.62 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951326 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18715543 104 - 22407834 ----------------UGGAGAACAUGGUGGUUACAAAUAACACUUCAACAUUUCAGGCCACAGUAAUGGUGGCGCUGGGCUAUUUCCUCAGUGCAGUUGCAGUUGCAACUGCAAUCCCG ----------------((..(((.(((((.((((....))))...........(((((((((.......))))).))))))))))))..)).(((((((((....)))))))))...... ( -34.80) >DroSec_CAF1 8390 104 - 1 ----------------AUAAGAACAAGGUGGUUACAAAUAACACUUUAACAUUUUUGACCACAGUAAUGGUGGCGCUGGGCUAUUUCCUCAGUGCAGUUGCAGUUGCAACUGCAAUCCCG ----------------...........(((((((.((((...........)))).)))))))......((.(((((((((.......)))))))).((((((((....))))))))))). ( -30.40) >DroSim_CAF1 6739 117 - 1 ---AAAUCGGUUUUAAAUGAGAACAUGGUGGUUGCAAAUAACACUUCAAUAUUUUUGGCCACAGUAAUGGUGGCGCUGGGCUAUUUCCUCAGUGCAGUUGCAGUUGCAACUGCAAUCCCG ---......(((((.....)))))..((.((((((......(((.((((.....))))...((....)))))((((((((.......))))))))((((((....)))))))))))))). ( -35.00) >DroEre_CAF1 8372 120 - 1 UCCAAAGUGAUUUAAAAUAAGAACCAGGUGGUUGCAAAUCACAGUUCAACAUUUUAGGCCACAGGAAUGUUGGCGCUGGGCUGUUUCCUCAGUGCAGUUGCAGUUGCAACUGCAAUCCCG ......(((((((.......(((((....)))).))))))))((((((((((((..(....)..))))))))).)))((((((......)))(((((((((....)))))))))..))). ( -38.41) >DroYak_CAF1 8390 120 - 1 UCCAAAGUGAUUCAACAUAAGAACAAGAUGGUUGCAACUCGCACUGCAACGUUUUUAGCCACAGGAAUGUUGGCGCUGGGCUAUUUCCUCAGUGCAGUUGCAGUUGCAACUGCAAUCCCG ......(.((((((.(((.........)))(((((((((.(((((.((((((((((......))))))))))((((((((.......)))))))))).)))))))))))))).)))).). ( -38.10) >consensus ___AAA__G_UU__A_AUAAGAACAAGGUGGUUGCAAAUAACACUUCAACAUUUUUGGCCACAGUAAUGGUGGCGCUGGGCUAUUUCCUCAGUGCAGUUGCAGUUGCAACUGCAAUCCCG ..........................((.((((........................((((((....)).))))((((((.......))))))((((((((....)))))))))))))). (-28.22 = -28.62 + 0.40)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:17:05 2006