
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,710,140 – 18,710,295 |
| Length | 155 |
| Max. P | 0.998690 |

| Location | 18,710,140 – 18,710,236 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.68 |
| Mean single sequence MFE | -24.48 |
| Consensus MFE | -13.73 |
| Energy contribution | -13.72 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.56 |
| SVM decision value | 3.05 |
| SVM RNA-class probability | 0.998251 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18710140 96 + 22407834 UAAAAAUAAA-------AUAUUAGCCCAGAUUAAAAAAGAAAAA-----------------UAAAUCUUAAAGAUUUGAAUAGCUCAGAUCACAACCAGACUGGUUGGGUUCAAGGCUAG ..........-------....(((((..((......((((....-----------------....))))...(((((((.....))))))).(((((.....)))))...))..))))). ( -20.90) >DroSec_CAF1 3084 113 + 1 UAAAAAUAAA-------AUAUUGGGCCUGAUUAAAAAACAAUAUGCAAUUCAAUCCUUUGGCAAAGUUUAAAGAUUUGAAUAGCUCAGAUCACAACCAGACUGGUUGGGUUCCAGGCUAA ..........-------......((((((...............((.((((((..((((((......))))))..)))))).))...((((.(((((.....))))))))).)))))).. ( -29.10) >DroSim_CAF1 2962 113 + 1 UAAAAAAAAA-------AUUUUGCACCUGAUUAAAAAACAAUAUGCCAUUCAAUCCUUUGGCAAAGUUUAAAAAUUUGAAUAGCUCAGAUCACAACCAGACUGGUUGGGUUCCAGGCUAA ........((-------(((((((....((((...................)))).....)))))))))...........(((((..((...(((((.....)))))...))..))))). ( -21.51) >DroYak_CAF1 2895 120 + 1 AAAUAAUAAAACAAGAAAUUUUGUGCCAGAUUAAAAUGGAAUAGGCAUAGCAAUCCUUCGACCAAAUUUAAAGAUUUAAAUAGCUAAGAUCGCAACCAGACCGGUUGCGUUCCAGGCUAA ..............(((((((((((((................))))))).)))..))).....................(((((..((.(((((((.....))))))).))..))))). ( -26.39) >consensus UAAAAAUAAA_______AUAUUGGGCCAGAUUAAAAAACAAUAUGCAAUUCAAUCCUUUGGCAAAGUUUAAAGAUUUGAAUAGCUCAGAUCACAACCAGACUGGUUGGGUUCCAGGCUAA ................................................................................(((((..((...(((((.....)))))...))..))))). (-13.73 = -13.72 + -0.00)



| Location | 18,710,140 – 18,710,236 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.68 |
| Mean single sequence MFE | -24.25 |
| Consensus MFE | -12.43 |
| Energy contribution | -12.67 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.650022 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18710140 96 - 22407834 CUAGCCUUGAACCCAACCAGUCUGGUUGUGAUCUGAGCUAUUCAAAUCUUUAAGAUUUA-----------------UUUUUCUUUUUUAAUCUGGGCUAAUAU-------UUUAUUUUUA .((((((.((...(((((.....))))).(((.((((...)))).)))...((((....-----------------....))))......)).))))))....-------.......... ( -17.70) >DroSec_CAF1 3084 113 - 1 UUAGCCUGGAACCCAACCAGUCUGGUUGUGAUCUGAGCUAUUCAAAUCUUUAAACUUUGCCAAAGGAUUGAAUUGCAUAUUGUUUUUUAAUCAGGCCCAAUAU-------UUUAUUUUUA .((((..(((.(.(((((.....))))).).)))..))))..................(((....(((((((..((.....))..))))))).))).......-------.......... ( -25.10) >DroSim_CAF1 2962 113 - 1 UUAGCCUGGAACCCAACCAGUCUGGUUGUGAUCUGAGCUAUUCAAAUUUUUAAACUUUGCCAAAGGAUUGAAUGGCAUAUUGUUUUUUAAUCAGGUGCAAAAU-------UUUUUUUUUA .((((..(((.(.(((((.....))))).).)))..))))...............((((((....(((((((..((.....))..)))))))..).)))))..-------.......... ( -26.30) >DroYak_CAF1 2895 120 - 1 UUAGCCUGGAACGCAACCGGUCUGGUUGCGAUCUUAGCUAUUUAAAUCUUUAAAUUUGGUCGAAGGAUUGCUAUGCCUAUUCCAUUUUAAUCUGGCACAAAAUUUCUUGUUUUAUUAUUU .((((..(((.(((((((.....))))))).)))..))))..((((.(...((((((.((.(..((((((..(((.......)))..))))))..))).))))))...).))))...... ( -27.90) >consensus UUAGCCUGGAACCCAACCAGUCUGGUUGUGAUCUGAGCUAUUCAAAUCUUUAAACUUUGCCAAAGGAUUGAAUUGCAUAUUCUUUUUUAAUCAGGCCCAAAAU_______UUUAUUUUUA .((((..(((.(.(((((.....))))).).)))..))))................................................................................ (-12.43 = -12.67 + 0.25)

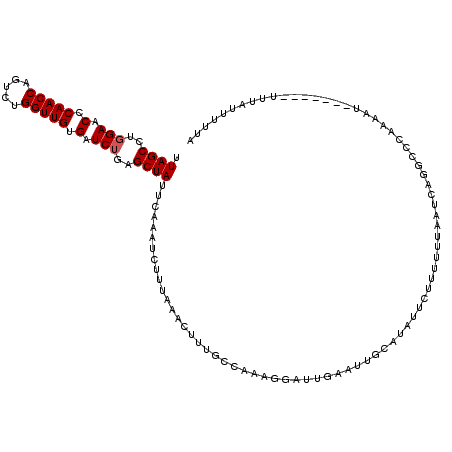

| Location | 18,710,196 – 18,710,295 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.23 |
| Mean single sequence MFE | -27.16 |
| Consensus MFE | -14.90 |
| Energy contribution | -14.94 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.55 |
| SVM decision value | 3.19 |
| SVM RNA-class probability | 0.998690 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18710196 99 + 22407834 UAGCUCAGAUCACAACCAGACUGGUUGGGUUCAAGGCUAGAUCUAUGUAU------GUAAAA---------------UUCGUCGUAGAAUCAAUGUAUAAUUCAUACAGAGAAGAGAAUG (((((..((((.(((((.....)))))))))...)))))..((((((.((------(.....---------------..))))))))).((..(((((.....)))))..))........ ( -23.70) >DroSec_CAF1 3157 99 + 1 UAGCUCAGAUCACAACCAGACUGGUUGGGUUCCAGGCUAAAUAUAUGUAU------GUCAAA---------------AUCUUCGUAAAAUCGAUGUAUAAUACAUACAAAAUACAGAAUG (((((..((...(((((.....)))))...))..)))))......(((((------((....---------------....(((......)))........)))))))............ ( -22.69) >DroSim_CAF1 3035 99 + 1 UAGCUCAGAUCACAACCAGACUGGUUGGGUUCCAGGCUAAAUCGAUGUAU------GUCAAA---------------UUCUUCGUAGAAUCGAUGUAUAAUACAUACAAAAUACAGAAUG (((((..((...(((((.....)))))...))..)))))......(((((------((....---------------....(((......)))........)))))))............ ( -22.69) >DroEre_CAF1 2983 120 + 1 UAGCUCAGAUCGCAACCAGACUGGUUGCGUUCCAAGCUAAAUCGUUGUAUGUAUAUGUAUAAUACAUGUAUGAAAGGUUCUUCGUAAAAUCUAUGUAUAAUACAUACAUAAUACCGGAUG (((((..((.(((((((.....))))))).))..)))))..(((...((((((..(((((.(((((((((((((......)))))).....))))))).)))))))))))....)))... ( -37.30) >DroYak_CAF1 2975 107 + 1 UAGCUAAGAUCGCAACCAGACCGGUUGCGUUCCAGGCUAAAUCUUUCUAU------GUAAAAUAAAUGUAAGUA-------UAGUAUAACCUAUGUAUAGUACCUACAUCCUGUAGGAUG (((((..((.(((((((.....))))))).))..)))))...........------...........(((.(((-------(.(((((.....))))).)))).)))((((....)))). ( -29.40) >consensus UAGCUCAGAUCACAACCAGACUGGUUGGGUUCCAGGCUAAAUCUAUGUAU______GUAAAA_______________UUCUUCGUAAAAUCGAUGUAUAAUACAUACAAAAUACAGAAUG (((((..((...(((((.....)))))...))..)))))......................................................(((((.....)))))............ (-14.90 = -14.94 + 0.04)



| Location | 18,710,196 – 18,710,295 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.23 |
| Mean single sequence MFE | -26.21 |
| Consensus MFE | -13.10 |
| Energy contribution | -13.50 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.50 |
| SVM decision value | 2.58 |
| SVM RNA-class probability | 0.995462 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18710196 99 - 22407834 CAUUCUCUUCUCUGUAUGAAUUAUACAUUGAUUCUACGACGAA---------------UUUUAC------AUACAUAGAUCUAGCCUUGAACCCAACCAGUCUGGUUGUGAUCUGAGCUA ........(((.((((((...........(((((......)))---------------))...)------))))).)))..((((...((.(.(((((.....))))).).))...)))) ( -18.34) >DroSec_CAF1 3157 99 - 1 CAUUCUGUAUUUUGUAUGUAUUAUACAUCGAUUUUACGAAGAU---------------UUUGAC------AUACAUAUAUUUAGCCUGGAACCCAACCAGUCUGGUUGUGAUCUGAGCUA ......((((..(((((((........(((......)))....---------------....))------))))).)))).((((..(((.(.(((((.....))))).).)))..)))) ( -22.79) >DroSim_CAF1 3035 99 - 1 CAUUCUGUAUUUUGUAUGUAUUAUACAUCGAUUCUACGAAGAA---------------UUUGAC------AUACAUCGAUUUAGCCUGGAACCCAACCAGUCUGGUUGUGAUCUGAGCUA ............(((((((...((.(.(((......))).).)---------------)...))------)))))......((((..(((.(.(((((.....))))).).)))..)))) ( -22.80) >DroEre_CAF1 2983 120 - 1 CAUCCGGUAUUAUGUAUGUAUUAUACAUAGAUUUUACGAAGAACCUUUCAUACAUGUAUUAUACAUAUACAUACAACGAUUUAGCUUGGAACGCAACCAGUCUGGUUGCGAUCUGAGCUA .(((..((((..(((((((((.((((((.(......)((((....))))....)))))).))))))))).))))...))).(((((..((.(((((((.....))))))).))..))))) ( -38.50) >DroYak_CAF1 2975 107 - 1 CAUCCUACAGGAUGUAGGUACUAUACAUAGGUUAUACUA-------UACUUACAUUUAUUUUAC------AUAGAAAGAUUUAGCCUGGAACGCAACCGGUCUGGUUGCGAUCUUAGCUA .........(((((((((((.......(((......)))-------))))))))))).......------...........((((..(((.(((((((.....))))))).)))..)))) ( -28.61) >consensus CAUUCUGUAUUAUGUAUGUAUUAUACAUAGAUUCUACGAAGAA_______________UUUUAC______AUACAUAGAUUUAGCCUGGAACCCAACCAGUCUGGUUGUGAUCUGAGCUA ............((((((...))))))......................................................((((..(((.(.(((((.....))))).).)))..)))) (-13.10 = -13.50 + 0.40)

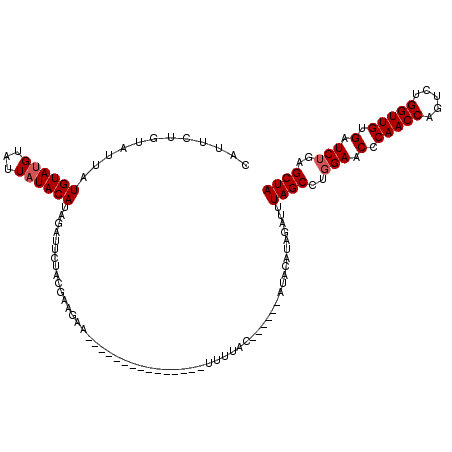
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:16:55 2006