
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,700,954 – 18,701,154 |
| Length | 200 |
| Max. P | 0.914632 |

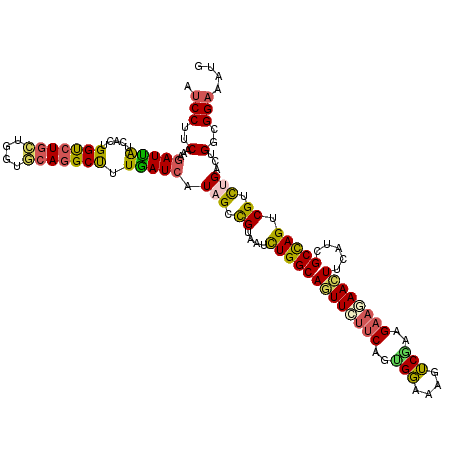
| Location | 18,700,954 – 18,701,074 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.67 |
| Mean single sequence MFE | -41.80 |
| Consensus MFE | -28.84 |
| Energy contribution | -28.32 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.41 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.914632 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18700954 120 - 22407834 AUCCUUCAAAGAUUAUUACUGGUCUGCUGGUGCAGGCUUUGAUCAUAGGCGUUAUCUGGCAGUUCUUCAGCGGAAAGUCGAAGAAGAACUUCAUCGCCAGUCGUCUGACUGGUGGAAAUG .(((.(((..(((((.....(((((((....))))))).))))).((((((....(((((((((((((..(((....)))..)))))))).....))))).))))))..))).))).... ( -44.20) >DroSec_CAF1 273 120 - 1 AUCCUUCAAAGAUUAUUACUGGUCUGCUGGUGCAGGCUUUGAUCAUAGGCGUUAUCUGGCAGUUCUUCAGCGGAAAGUCGAAGAAGAACUUCAUCGCUAGUCGUCUGACCGGCGGAAAUG .(((.((...(((((.....(((((((....))))))).))))).((((((....(((((((((((((..(((....)))..)))))))).....))))).))))))...)).))).... ( -41.40) >DroEre_CAF1 271 118 - 1 AUCCUUCAAAGAUUGUCACUGGUCUGCUGGUGCAGGCUAUAAUCAUAGCCGUAAUCUGGCAGUUCUUCAGUGGAAAGUCGAAGAAUAAAUUCAUCGCCAGUCGUCUGACUGGGGGAAA-- .((((.....((((.(((((((.((((..(....((((((....)))))).....)..))))....)))))))..))))((.(((....))).)).((((((....))))))))))..-- ( -42.40) >DroWil_CAF1 279 120 - 1 AUCCCACGAAAAUUGUGAGUGGUCUCAUUGUUCAAGCUCUGAUCAUUGCCGUAAUUUGGCAAUUUUUCAGUGUGAAAACAAAGAAAAAUUUUAUUGCCAGUCGUCUGGGUGGUGGCAAUG .((((((.((((((.((((....))))(((((...((.((((..(((((((.....)))))))...)))).))...))))).....))))))....((((....)))))))).))..... ( -33.40) >DroYak_CAF1 273 120 - 1 AUCCUUCAAAGAUUGUCACUGGUCUGCUGGUGCAGGCUUUAAUCAUAGGCGUAAUCUGGCAGUUCUUCAGUGGAAAGUCGAAGAAGAACUUCAUCGCCAGUCGUCUGACAGGGGGAAAUG .((((((...(((((.....(((((((....))))))).))))).((((((....(((((((((((((..(((....)))..)))))))).....))))).))))))...)))))).... ( -45.50) >DroAna_CAF1 273 120 - 1 AUCCGGCCAAGAUCAUCAGUGGCCUGAUUGUCCAGGCCCUGAUCAUCGCUGUGAUCUGGCAGUUUUUCAGUGGAAAGUCCAAGUCCAACUUUAUAGCUAGCCGUUUGACUGGCGGAAAUG .((((.(((.(((.(((((.((((((......))))))))))).)))...((.(..((((((((....((((((.........))).)))....)))).))))..).))))))))).... ( -43.90) >consensus AUCCUUCAAAGAUUAUCACUGGUCUGCUGGUGCAGGCUUUGAUCAUAGCCGUAAUCUGGCAGUUCUUCAGUGGAAAGUCGAAGAAGAACUUCAUCGCCAGUCGUCUGACUGGCGGAAAUG .(((..(...(((((.....(((((((....))))))).))))).(((.((....(((((((((((((..(((....)))..)))))))).....))))).)).)))...)..))).... (-28.84 = -28.32 + -0.52)


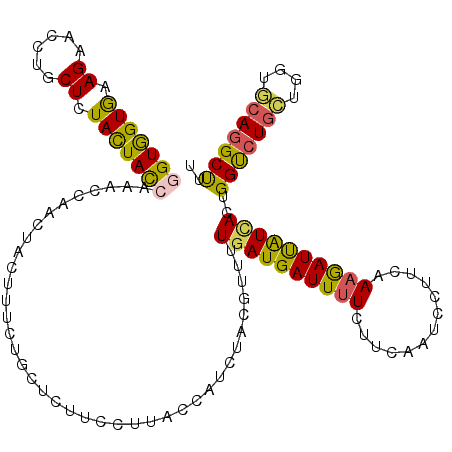
| Location | 18,701,034 – 18,701,154 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.89 |
| Mean single sequence MFE | -27.39 |
| Consensus MFE | -19.78 |
| Energy contribution | -19.20 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.556118 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18701034 120 - 22407834 GGUGGUGAAGAACCUGCUCUAUUACCAAACAAACUACUUUUUGCUCUUCAUUACCAUCUACGUUUUGAUGAUUUUCUUCAAUCCUUCAAAGAUUAUUACUGGUCUGCUGGUGCAGGCUUU (((((((((((....((.........................))))))))))))).......((((((.((((......))))..)))))).........(((((((....))))))).. ( -28.11) >DroSim_CAF1 353 120 - 1 GGUGGUGAAGAACCUGCUCUACUAUCAAACAAAUUACUUUUUGCUCUUCCUUACCAUCUACGCUUUGAUGAUUUUCUGCAAUCCUUCAAAGAUUAUUACUGGUCUGCUGGUGCAGGCUUU ((((((((.(((...((.........................))..))).))))))))....((((((.((((......))))..)))))).........(((((((....))))))).. ( -28.41) >DroEre_CAF1 349 120 - 1 GGUGGUGAAGAACCUGCUGUACUACCAAACCAACUACUUUCUGCUCUUCCUUACCAUCUACGUUUUGAUGAUUUUCUUCAAUCCUUCAAAGAUUGUCACUGGUCUGCUGGUGCAGGCUAU ((((((((.(((...((.........................))..))).)))))))).......(((..(((((.............)))))..))).((((((((....)))))))). ( -27.63) >DroWil_CAF1 359 120 - 1 UGUGGUAAAGAAUUUACUUUACUACCAAACCAAUUAUUUCCUACUCUUCCUUACAAUCUAUGCCCUGAUGAUUUUCUUCAAUCCCACGAAAAUUGUGAGUGGUCUCAUUGUUCAAGCUCU .(((((((((......)))))))))........................((((((((....(((((.(..((((((...........))))))..).)).)))...)))))..))).... ( -22.60) >DroYak_CAF1 353 120 - 1 GGUAGUGAAGAAUUUGCUGUACUACCAAACCAACUACUUUCUGCUCUUCCUUACGAUUUACGUUUUGAUGAUUUUCUUCAAUCCUUCAAAGAUUGUCACUGGUCUGCUGGUGCAGGCUUU (((((((.((......)).)))))))............................(((.....((((((.((((......))))..))))))...)))...(((((((....))))))).. ( -26.80) >DroAna_CAF1 353 120 - 1 AGUGGUGAAGAACUUGCUCUACUACCAAACCAAUUACUUUUUGCUCUUCCUGACCAUCUACGUCCUGAUGAUUGUCUGCAAUCCGGCCAAGAUCAUCAGUGGCCUGAUUGUCCAGGCCCU .(((((.(.(((...((.........................))..))).).))))).......(((((((((....((......))...))))))))).((((((......)))))).. ( -30.81) >consensus GGUGGUGAAGAACCUGCUCUACUACCAAACCAACUACUUUCUGCUCUUCCUUACCAUCUACGUUUUGAUGAUUUUCUUCAAUCCUUCAAAGAUUAUCACUGGUCUGCUGGUGCAGGCUUU (((((((.((......)).))))))).......................................((((((((((.............))))))))))..(((((((....))))))).. (-19.78 = -19.20 + -0.58)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:16:50 2006