
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,698,878 – 18,698,972 |
| Length | 94 |
| Max. P | 0.773177 |

| Location | 18,698,878 – 18,698,972 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 81.36 |
| Mean single sequence MFE | -25.83 |
| Consensus MFE | -15.64 |
| Energy contribution | -16.83 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.61 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
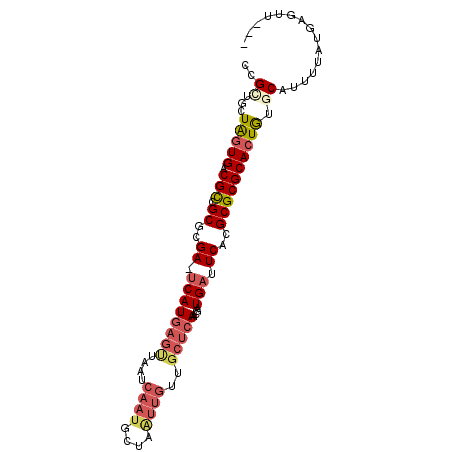
>2L_DroMel_CAF1 18698878 94 + 22407834 CCGCUCCUAGUGACGCCGCGCGA-UCAUGAGCUUAUCAAUGCUAAUUGUUGCUCAACGAUGAUUCACGCGCGCACUGUGCAUUUUAUGAUUUUUU ..((...(((((.(((.(((.((-((((((((....((((....))))..)))))....))).)).))))))))))).))............... ( -28.20) >DroVir_CAF1 4608 74 + 1 ACGUUAUUGUUGGCGC-GCCUGACACAUGUGUUAAU----AUUGGUAAU-----AACGAUGAUUCACGCGCGCACUA-GCAUUUU---------- .......(((((((((-((.(((..(((.(((((.(----(....)).)-----)))))))..))).)))))).).)-)))....---------- ( -20.40) >DroSec_CAF1 3944 91 + 1 CCGCUGCUAGUGGCGCCGCGCGA-UCAUGAGUUAAUCAAUGCUAAUUGUUGCUCAACGAUGAUUCACGCGCGCACUGUGCAUUUUAUGAGUU--- ....((((((((.(((.(((.((-((((((((....((((....))))..)))))....))).)).))))))))))).)))...........--- ( -27.90) >DroSim_CAF1 3968 91 + 1 CCGCUGCUAGUGGCGCCGCGCGA-UCAUGAGUUAAUCAAUGCUAAUUGUUGCUCAACGAUGAUUCACGCGCGCACUGUGCAUUUUAUGAGUU--- ....((((((((.(((.(((.((-((((((((....((((....))))..)))))....))).)).))))))))))).)))...........--- ( -27.90) >DroEre_CAF1 3896 91 + 1 CCGCUGCUAGUGACGUCGCACGA-UCAUGAGUUAAUCAAUGUUAAUUGUUGCUCAACGAUGAUUCACGCGCGCACUGGCCAUUUUAUGAGUU--- .....(((((((.((.(((..((-((((((((....((((....))))..)))))....)))))...)))))))))))).............--- ( -27.10) >DroYak_CAF1 3912 89 + 1 CCGCUGCUAGUGACGUCGCACGA-UCAUGAGUUAAUCAAUGUUAGUUGUUGCUCAACGAUGAUUCACGCGCGCACUG--CAUUUUAUGAGUU--- ....(((.((((.((.(((..((-((((((((....((((....))))..)))))....)))))...))))))))))--))...........--- ( -23.50) >consensus CCGCUGCUAGUGACGCCGCGCGA_UCAUGAGUUAAUCAAUGCUAAUUGUUGCUCAACGAUGAUUCACGCGCGCACUGUGCAUUUUAUGAGUU___ ..((...(((((.(((.((..((.((((((((....((((....))))..)))))....))).))..)))))))))).))............... (-15.64 = -16.83 + 1.20)

| Location | 18,698,878 – 18,698,972 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 81.36 |
| Mean single sequence MFE | -24.62 |
| Consensus MFE | -18.08 |
| Energy contribution | -19.22 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.773177 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18698878 94 - 22407834 AAAAAAUCAUAAAAUGCACAGUGCGCGCGUGAAUCAUCGUUGAGCAACAAUUAGCAUUGAUAAGCUCAUGA-UCGCGCGGCGUCACUAGGAGCGG ..............(((.((((((((((((((.(((....(((((..((((....))))....))))))))-)))))).))).)))).)..))). ( -28.20) >DroVir_CAF1 4608 74 - 1 ----------AAAAUGC-UAGUGCGCGCGUGAAUCAUCGUU-----AUUACCAAU----AUUAACACAUGUGUCAGGC-GCGCCAACAAUAACGU ----------.......-..(.((((((.(((..(((.(((-----(........----..))))..)))..))).))-)))))........... ( -19.40) >DroSec_CAF1 3944 91 - 1 ---AACUCAUAAAAUGCACAGUGCGCGCGUGAAUCAUCGUUGAGCAACAAUUAGCAUUGAUUAACUCAUGA-UCGCGCGGCGCCACUAGCAGCGG ---...........(((..(((((((((((((.((((.(((((....((((....)))).)))))..))))-)))))).))).)))).))).... ( -26.90) >DroSim_CAF1 3968 91 - 1 ---AACUCAUAAAAUGCACAGUGCGCGCGUGAAUCAUCGUUGAGCAACAAUUAGCAUUGAUUAACUCAUGA-UCGCGCGGCGCCACUAGCAGCGG ---...........(((..(((((((((((((.((((.(((((....((((....)))).)))))..))))-)))))).))).)))).))).... ( -26.90) >DroEre_CAF1 3896 91 - 1 ---AACUCAUAAAAUGGCCAGUGCGCGCGUGAAUCAUCGUUGAGCAACAAUUAACAUUGAUUAACUCAUGA-UCGUGCGACGUCACUAGCAGCGG ---.............((.(((((((((((((.((((.(((((....((((....)))).)))))..))))-))))))).)).)))).))..... ( -23.00) >DroYak_CAF1 3912 89 - 1 ---AACUCAUAAAAUG--CAGUGCGCGCGUGAAUCAUCGUUGAGCAACAACUAACAUUGAUUAACUCAUGA-UCGUGCGACGUCACUAGCAGCGG ---...........((--((((((((((((((.((((.(((((.(((.........))).)))))..))))-))))))).)).)))).))).... ( -23.30) >consensus ___AACUCAUAAAAUGCACAGUGCGCGCGUGAAUCAUCGUUGAGCAACAAUUAACAUUGAUUAACUCAUGA_UCGCGCGGCGCCACUAGCAGCGG ...................(((((((((((((.((((.(((((.(((.........))).)))))..)))).))))))).)).))))........ (-18.08 = -19.22 + 1.14)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:16:48 2006