| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,686,407 – 18,686,516 |
| Length | 109 |
| Max. P | 0.850674 |

| Location | 18,686,407 – 18,686,516 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 80.61 |
| Mean single sequence MFE | -41.97 |
| Consensus MFE | -30.12 |
| Energy contribution | -31.43 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.850674 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
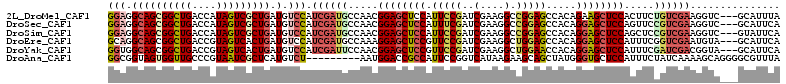
>2L_DroMel_CAF1 18686407 109 - 22407834 GGAGGCAGCGGCUGACCAUAGUCGCUGAUGUCCAUCGAUGCCAACGGAGCUCCAUUCCGAUCGAAGGCCGGAGCCACAGAAGCUCCACUUCUGUCGAAGGUC---GCAUUUA (((.((((((((((....))))))))).).)))...(((((...(((((.....)))))......((((....(.(((((((.....))))))).)..))))---))))).. ( -41.50) >DroSec_CAF1 13385 109 - 1 GGAGGCAGCGGCUGACCAUAGUCGCUGAUGUCCAUCGAUGCCAACGGAGCUCCAUUUCGAUCGAAGGCCGGAGCCACAGGAGCUCCAGUUCCGUCGAAGGUC---GCAUUCA (((.((((((((((....))))))))...(.((.((((((..(((((((((((..(((....)))(((....)))...)))))))).))).)))))).)).)---)).))). ( -47.40) >DroSim_CAF1 11656 109 - 1 GGAGGCAGCGGCUGACCAUAGUCGCUGAUGUCCAUCGAUGCCAACGGAGCUCCAUUCCGAUCGAAGGCCGGAGCCACAGGAGCUCCAGCUCCGUCGAAGGUC---GUAUUCA .(((.(((((((((....)))))))))(((.((.((((((.....((((((((.(((.....)))(((....)))...)))))))).....)))))).)).)---)).))). ( -46.60) >DroEre_CAF1 11449 109 - 1 GCAGGCAGCGGCUGACCGUAGUCACUGAUGUCCAUCGAUGCCAAAGGAGCUCCGUUCCGAUCGAAGGCUGGAGCCACAGGAGCUCCAUUUCGGUCGAAUGUA---GCAUUCA ...((((.(((..(((..(((...)))..)))..))).))))...((((((.(((((.((((((((..(((((((....).))))))))))))))))))).)---)).))). ( -40.40) >DroYak_CAF1 11661 109 - 1 GGUGGCAGCGGCUGACCGUAGUCACUGAUGUCCAUCGAUUCCAACGGAGCUCCGUUCCGAUCGAAGGCUGGAACCACAGGAGCUCCAUUUCGAUCGACGGUA---GCAUUCA ((((((.((((....)))).))))))(((((((.((((((..((.((((((((((((((..(....).))))))....)))))))).))..)))))).))..---))))).. ( -46.60) >DroAna_CAF1 11566 103 - 1 GGCGGUAGUGGUUGCCCGUAAUCGCUCAUGUCU---------AAUGGACCGCCAUUCCGGUCAUAAGAAGCAGCUAUGGGUGCUCCAUUUCUAUCAAAAGCAGGGGCGUUUA (..((.(((((..(((((((...(((....(((---------.(((.((((......))))))).)))...))))))))))...))))).))..)....((....))..... ( -29.30) >consensus GGAGGCAGCGGCUGACCAUAGUCGCUGAUGUCCAUCGAUGCCAACGGAGCUCCAUUCCGAUCGAAGGCCGGAGCCACAGGAGCUCCAUUUCCGUCGAAGGUA___GCAUUCA (((.((((((((((....))))))))).).))).((((((.....((((((((.(((((..(....).))))).....)))))))).....))))))............... (-30.12 = -31.43 + 1.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:16:40 2006