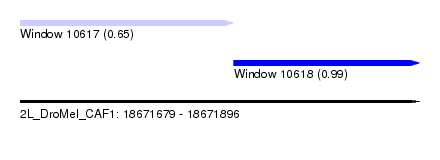
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,671,679 – 18,671,896 |
| Length | 217 |
| Max. P | 0.989950 |

| Location | 18,671,679 – 18,671,795 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 74.76 |
| Mean single sequence MFE | -26.45 |
| Consensus MFE | -12.29 |
| Energy contribution | -13.02 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.649696 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18671679 116 + 22407834 UUCGAAUGCUUUCCCUAAGCUUUCCCACCUCCAACUCAUACAUGAGUCCGGAGGAAUACUGCACC---UUUUCAUCCUGCACUGCUUGUACCUCCAGGUCUUGUAGCUCUCUUUUUAUC ...(((.((((.....)))).)))...(((((.(((((....)))))..)))))...........---........(((((..(((((......)))))..)))))............. ( -27.70) >DroSec_CAF1 24494 116 + 1 UUCUGAUGCUUUCCCUAAGCUUUCCCACCUCCAACUCAUACAUGAGUCCGGAGGAAUACUGCACC---UUUUCAUCCUGCACUGCUUGUACCUCCAGGUCAUGUAGUUCUUUUUUUAUC ...(((.((((.....)))).......(((((.(((((....)))))..)))))...........---...)))..(((((..(((((......)))))..)))))............. ( -26.20) >DroSim_CAF1 25806 116 + 1 UUCUGAUGCUUUCCCUAAGCUUUCCCACCUCCAACUCAUACAUGAGUCCGGAGGAAUACUGCACC---UUUUCAUCCUGCACUGCUUGAACCUCCAGGUCAUGUAGUUCUUUCUUUAUC ...(((.((((.....)))).......(((((.(((((....)))))..)))))...........---...)))..(((((..(((((......)))))..)))))............. ( -26.70) >DroEre_CAF1 24826 116 + 1 UUCUAAUGCUAUUCCUAAGCUCUUCCACCUCCAGCGAAUACAUGAGCUGGGAGGAAUGCUGCACC---GAUUCCUCCUGCAAUGCUUGUUCUUCCAGGUCGUGCAGUUCUUUUCUUAUC .....(((.(((((....(((...........)))))))))))((((((((((((((.(......---))))))))))(((..(((((......)))))..)))))))).......... ( -28.40) >DroYak_CAF1 24710 116 + 1 UUUCAAUGCUUUGCCUAAGCUUUUCCACCUCCAACGAAUACAUGAGUUCUGAGGAAUGCUGCACC---GAUUCAUCCUGCAAUGCUUGUACCUCCAGGUCGUGGAGUUCUUUUUUUACC .......((((.....))))..((((((..((...((((......)))).(((((..((((((..---((....)).))))..))...).))))..))..))))))............. ( -23.20) >DroAna_CAF1 20568 116 + 1 CUCCGAUCAUCUGCUGCAGCCUCUGAAUCUCGUACAAAUACACCGCCUCGGAGUGGAGGUGCACGGCAUUGUCAACGUCACAUGUC---UCCUGCAGGCUGGAUAGUUCUUCACAUAGC ....((....(((.(.(((((((.((.....((((...(((.(((...))).)))...))))..(((((((.......)).)))))---))..).)))))).))))....))....... ( -26.50) >consensus UUCUAAUGCUUUCCCUAAGCUUUCCCACCUCCAACUAAUACAUGAGUCCGGAGGAAUACUGCACC___UUUUCAUCCUGCAAUGCUUGUACCUCCAGGUCGUGUAGUUCUUUUUUUAUC ..................((.......(((((.................)))))......))..............(((((..(((((......)))))..)))))............. (-12.29 = -13.02 + 0.72)


| Location | 18,671,795 – 18,671,896 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.92 |
| Mean single sequence MFE | -34.50 |
| Consensus MFE | -16.75 |
| Energy contribution | -16.87 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.42 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.49 |
| SVM decision value | 2.19 |
| SVM RNA-class probability | 0.989950 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18671795 101 + 22407834 UCCACAUUUCUUUCGUUCAUCCGGGUCAGACGCUGAUCCGUACAGCUUUUAAACCAGUUGAUUUGAUAUCGCGUUUCAAUCGGGUUCAGGGCUGCGGAAGA------------------- ........(((((((......((((((((...))))))))..(((((((.((.((.(((((..((......))..))))).)).)).))))))))))))))------------------- ( -32.40) >DroSec_CAF1 24610 101 + 1 UCCACAUUUCUUCCAUUCAUCCGGGUCAGACGCUGAUCCGUACAGCUUCUAAACCAGUUGAUUUGAUAUCGCGUUUCAAUCGGGUUCAUCGCUGCGGAAGA------------------- ........((((((.......((((((((...))))))))..((((.......((.(((((..((......))..))))).)).......)))).))))))------------------- ( -31.04) >DroSim_CAF1 25922 101 + 1 UCCACAUUUCUUCCAUUCAUCCGGGUCAGACGCUGAUCCGUACAGCUCCUAAACCAGUUGAUUUGAUAUCGCGUUUCAAUCGGGUUCAUCGCUGCGGAAGA------------------- ........((((((.......((((((((...))))))))..((((.......((.(((((..((......))..))))).)).......)))).))))))------------------- ( -31.04) >DroEre_CAF1 24942 101 + 1 UCAUCCUCUCUUCCGUUUAGCCGGCAGAGACGCUGAUCCGUGCAGCUUCUAAUCCAAUUGAUUUGAAAUCGGAUUUCAAUUGGAUCUAGGGCUGCGGAAGG------------------- (((.(.(((((.(((......))).))))).).)))(((..((((((.(((((((((((((((((....))))..)))))))))).))))))))))))...------------------- ( -38.60) >DroYak_CAF1 24826 101 + 1 UCCUCCUCUUUUCCGCUUAUCCGGAAGAGACGCUGAUCCGUACAGCUUCUAAUCCAAUUGAUUUGGUAUCGCAUUUCAAUCGGAUCCUGGGCUGCGGAAGG------------------- .(((..(((((((((......)))))))))......((((..((((((...((((.(((((..((......))..))))).))))...)))))))))))))------------------- ( -34.00) >DroAna_CAF1 20684 120 + 1 UCAUCCUCCUGGUGGUCCAUGCGCAGGACUCGCUGGUCAGCACAGCUCCUGAUCCAAUUCACUUGGUAGCGCUCCUCGAUGGGGUCAAGUGCUGUGGAAGGCAGAAAUAAGGCCAUUAUA ...(((..(..(((((((.......)))).)))..).((((((.((((((((......)))...)).)))(((((.....)))))...)))))).))).(((.........)))...... ( -39.90) >consensus UCCACAUCUCUUCCGUUCAUCCGGGUCAGACGCUGAUCCGUACAGCUUCUAAACCAAUUGAUUUGAUAUCGCGUUUCAAUCGGGUCCAGGGCUGCGGAAGA___________________ ........((((((((...........(((.((((.......)))).)))...((.(((((..((......))..))))).))..........))))))))................... (-16.75 = -16.87 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:16:36 2006