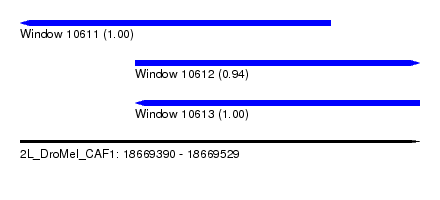
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,669,390 – 18,669,529 |
| Length | 139 |
| Max. P | 0.999840 |

| Location | 18,669,390 – 18,669,498 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.67 |
| Mean single sequence MFE | -34.24 |
| Consensus MFE | -21.21 |
| Energy contribution | -21.56 |
| Covariance contribution | 0.35 |
| Combinations/Pair | 1.16 |
| Mean z-score | -4.51 |
| Structure conservation index | 0.62 |
| SVM decision value | 2.98 |
| SVM RNA-class probability | 0.997986 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18669390 108 - 22407834 AAGUCCGAAAGGAUGUCGGAAAAAAAAUCAGAGAGGAA------------UUCCGAAAAUAAGAUAGGACGAAAAUCUGAAAGGCUUUCUGAGAGACGGUCUGAAAGAAAGUCCCGAAAU ..((((....)))).((((........(((((......------------.(((............)))......)))))..((((((((...((.....))...))))))))))))... ( -29.52) >DroSec_CAF1 22094 120 - 1 AAGUCCGAAAGGAAGUCCGAGAGAAAGUCAGAGAGGAAGUCCGCGAGGAUUUCCGAAAAUAAGAUAGCACGAAAAUCUGAAAGGCUUUCUGAGAGAAGGUCUGAAAGGAAGUCCCGAAAU ..........(((..(((.........(((((..((((((((....))))))))........(....).......))))).((((((((.....))))))))....)))..)))...... ( -36.50) >DroSim_CAF1 23368 120 - 1 AAGUCCGAAAGGAAGUCCGAGAGAAAGUCAGAGAGGAAGUCCGCGAGGAUUUCAGAAAAUAAGAUAGCACGAAAAUCUGAAAGGCUUUCUGAGAGAAGGUCUGAAAGGAAGUCCCGAAAU ..........(((..(((.........(((((...(((((((....))))))).........(....).......))))).((((((((.....))))))))....)))..)))...... ( -33.00) >DroEre_CAF1 22378 120 - 1 AGUUCUGAGAGGAAGUCGCAGAGGAAGUCGCAGAGGAAGUCGCAGAGGAAGUCUGAUAAAAUGACAGGACGAAAGUCUGAAAGGCUUUCUGAGAGACGAUCUGAAAGAAAGUCCAGAAAU ...((((.((.....)).))))(((..((.((((....(((.(..(((((((((............((((....))))...)))))))))..).)))..))))...))...)))...... ( -35.76) >DroYak_CAF1 22275 120 - 1 AAGUCUGAGAGGAAGACCGAGAGGAAGUCAGAAAGGAAGUCCGAGAGGAAGUCUGAUAGAAUGAUAGGACGAAAGUCUGAAAGGCUUUCUGAAAGACGGUCUGAAAGAAAGUCCAGAAAU ..((((...((((((.((........((((....(((..(((....)))..))).......)))).((((....))))....))))))))...))))((.((.......)).))...... ( -36.40) >consensus AAGUCCGAAAGGAAGUCCGAGAGAAAGUCAGAGAGGAAGUCCGAGAGGAAUUCCGAAAAUAAGAUAGGACGAAAAUCUGAAAGGCUUUCUGAGAGACGGUCUGAAAGAAAGUCCCGAAAU ...(((....)))..............(((((..((((.(((....))).)))).....................)))))..((((((((...((.....))...))))))))....... (-21.21 = -21.56 + 0.35)


| Location | 18,669,430 – 18,669,529 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.76 |
| Mean single sequence MFE | -20.39 |
| Consensus MFE | -4.96 |
| Energy contribution | -5.99 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.24 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.938381 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18669430 99 + 22407834 UCAGAUUUUCGUCCUAUCUUAUUUUCGGAA------------UUCCUCUCUGAUUUUUUUUCCGACAUCCUUUCGGACUUUCUCUCGGAAUU---------UCGAACCGACUUCCUGGAC ..........((((.........(((((((------------((((.....((.......(((((.......)))))...))....))))))---------)))))..........)))) ( -21.41) >DroSec_CAF1 22134 111 + 1 UCAGAUUUUCGUGCUAUCUUAUUUUCGGAAAUCCUCGCGGACUUCCUCUCUGACUUUCUCUCGGACUUCCUUUCGGACUUUCUCUCGGAAUU---------UCGAACCGACUUCCUGGAC (((((.....(......)........((((.(((....))).))))..))))).....(((.(((..((..((((((.((((....)))).)---------)))))..))..))).))). ( -23.90) >DroSim_CAF1 23408 111 + 1 UCAGAUUUUCGUGCUAUCUUAUUUUCUGAAAUCCUCGCGGACUUCCUCUCUGACUUUCUCUCGGACUUCCUUUCGGACUUUCUCUCGGAAUU---------UCGAACCGACUUCCUGGAC (((((.....(......).........(((.(((....))).)))...))))).....(((.(((..((..((((((.((((....)))).)---------)))))..))..))).))). ( -19.90) >DroEre_CAF1 22418 108 + 1 UCAGACUUUCGUCCUGUCAUUUUAUCAGACUUCCUCUGCGACUUCCUCUGCGACUUCCUCUGCGACUUCCUCUCAGAACUUCUG---GA---------UAUUCUUACCGACCUCCUGGAC (((((.....(((..(((.........))).......(((........))))))....((((.((.....)).))))...))))---).---------........(((......))).. ( -14.30) >DroYak_CAF1 22315 120 + 1 UCAGACUUUCGUCCUAUCAUUCUAUCAGACUUCCUCUCGGACUUCCUUUCUGACUUCCUCUCGGUCUUCCUCUCAGACUUUCUCUCAGAAAUUUGAGAUAUUCUUACCGACCUCCUGGAU (((((.....((((........................))))......)))))..(((..(((((.....(((((((.((((.....))))))))))).......)))))......))). ( -22.46) >consensus UCAGAUUUUCGUCCUAUCUUAUUUUCAGAAAUCCUCGCGGACUUCCUCUCUGACUUUCUCUCGGACUUCCUUUCGGACUUUCUCUCGGAAUU_________UCGAACCGACUUCCUGGAC ((((....(((................(((.(((....))).)))...(((((...................)))))..............................)))....)))).. ( -4.96 = -5.99 + 1.03)

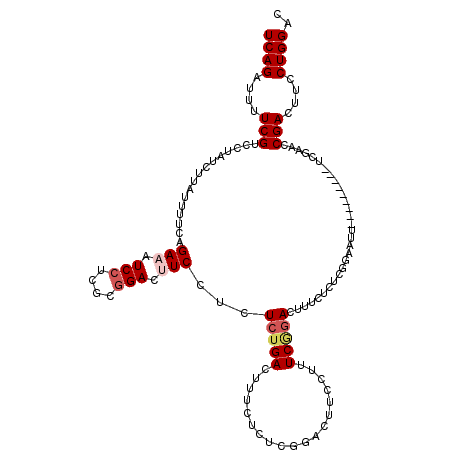
| Location | 18,669,430 – 18,669,529 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.76 |
| Mean single sequence MFE | -30.54 |
| Consensus MFE | -17.41 |
| Energy contribution | -17.84 |
| Covariance contribution | 0.43 |
| Combinations/Pair | 1.18 |
| Mean z-score | -3.49 |
| Structure conservation index | 0.57 |
| SVM decision value | 4.22 |
| SVM RNA-class probability | 0.999840 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
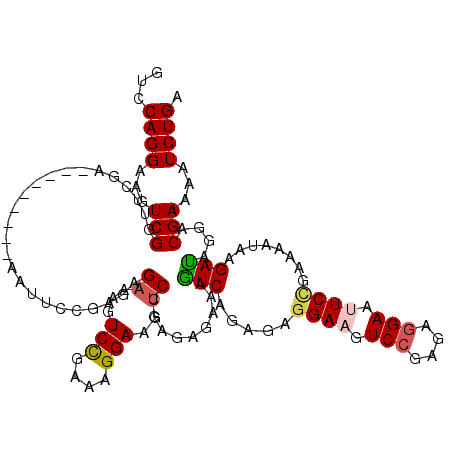
>2L_DroMel_CAF1 18669430 99 - 22407834 GUCCAGGAAGUCGGUUCGA---------AAUUCCGAGAGAAAGUCCGAAAGGAUGUCGGAAAAAAAAUCAGAGAGGAA------------UUCCGAAAAUAAGAUAGGACGAAAAUCUGA ((((.....(((..((((.---------((((((........((((....)))).((.((.......)).))..))))------------)).)))).....))).)))).......... ( -23.50) >DroSec_CAF1 22134 111 - 1 GUCCAGGAAGUCGGUUCGA---------AAUUCCGAGAGAAAGUCCGAAAGGAAGUCCGAGAGAAAGUCAGAGAGGAAGUCCGCGAGGAUUUCCGAAAAUAAGAUAGCACGAAAAUCUGA ...((((...(((((((((---------..((((..(.((...(((....)))..)))..).)))..)).....((((((((....))))))))........))..)).)))...)))). ( -34.90) >DroSim_CAF1 23408 111 - 1 GUCCAGGAAGUCGGUUCGA---------AAUUCCGAGAGAAAGUCCGAAAGGAAGUCCGAGAGAAAGUCAGAGAGGAAGUCCGCGAGGAUUUCAGAAAAUAAGAUAGCACGAAAAUCUGA ...((((...(((((((((---------..((((..(.((...(((....)))..)))..).)))..))......(((((((....))))))).........))..)).)))...)))). ( -31.40) >DroEre_CAF1 22418 108 - 1 GUCCAGGAGGUCGGUAAGAAUA---------UC---CAGAAGUUCUGAGAGGAAGUCGCAGAGGAAGUCGCAGAGGAAGUCGCAGAGGAAGUCUGAUAAAAUGACAGGACGAAAGUCUGA .(((.(((..((.....))...---------))---).((..((((....)))).)).....))).((((....(((..((......))..))).......)))).((((....)))).. ( -26.80) >DroYak_CAF1 22315 120 - 1 AUCCAGGAGGUCGGUAAGAAUAUCUCAAAUUUCUGAGAGAAAGUCUGAGAGGAAGACCGAGAGGAAGUCAGAAAGGAAGUCCGAGAGGAAGUCUGAUAGAAUGAUAGGACGAAAGUCUGA .(((....((((..........(((((......))))).....(((....))).))))....))).((((....(((..(((....)))..))).......)))).((((....)))).. ( -36.10) >consensus GUCCAGGAAGUCGGUUCGA_________AAUUCCGAGAGAAAGUCCGAAAGGAAGUCCGAGAGAAAGUCAGAGAGGAAGUCCGAGAGGAAUUCCGAAAAUAAGAUAGGACGAAAAUCUGA ...((((...(((.........................((...(((....)))..)).........(((.....((((.(((....))).))))........)))....)))...)))). (-17.41 = -17.84 + 0.43)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:16:32 2006