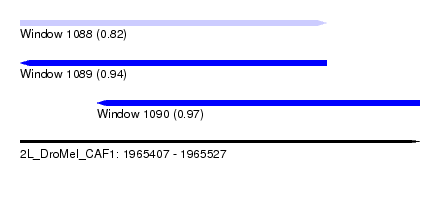
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,965,407 – 1,965,527 |
| Length | 120 |
| Max. P | 0.970535 |

| Location | 1,965,407 – 1,965,499 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.80 |
| Mean single sequence MFE | -35.18 |
| Consensus MFE | -28.62 |
| Energy contribution | -28.62 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.815921 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1965407 92 + 22407834 -----------------AGCAACGGAUUCUCGUUGGACCCUUUAGCGCAUUGGAUAGCGCGUUGGACUUCUAAUCCAAAGGUGGCGGGUUCGAUUCCCGCAAGGGUUGA----------- -----------------..((((((....))))))(((((((..((((........)))).(((((.......))))).....(((((.......))))))))))))..----------- ( -34.20) >DroSec_CAF1 7870 92 + 1 -----------------AGCAGCGGAUUCUCGUUGGACCCUUUAGCGCAUUGGAUAGCGCGUUGGACUUCUAAUCCAAAGGUGGCGGGUUCGAUUCCCGCAAGGGUUGA----------- -----------------..((((((....))))))(((((((..((((........)))).(((((.......))))).....(((((.......))))))))))))..----------- ( -34.20) >DroSim_CAF1 12009 92 + 1 -----------------AGCAGCGGAUUCUCGUUGGACCCUUUAGCGCAUUGGAUAGCGCGUUGGACUUCUAAUCCAAAGGUGGCGGGUUCGAUUCCCGCAAGGGUUGA----------- -----------------..((((((....))))))(((((((..((((........)))).(((((.......))))).....(((((.......))))))))))))..----------- ( -34.20) >DroEre_CAF1 17042 103 + 1 -----------------AUCCGGCGAAUCUCGUAGGACCCUUUAGCGCAUUGGAUAGCGCGUUGGACUUCUAAUCCAAAGGUGGCGGGUUCGAUUCCCGCAAGGGUUGAUUGCCAUUAGC -----------------....(((.((((.....((((((....((((........)))).(((((.......))))).......))))))....(((....)))..)))))))...... ( -35.70) >DroYak_CAF1 14896 103 + 1 -----------------AUGCGGCGAUUCGCGUAGGACCCUUUAGCGCAUUGGAUAGCGCGUUGGACUUCUAAUCCAAAGGUGGCGGGUUCGAUUCCCGCAAGGGUUGAUUGCCACUAGA -----------------....(((((((.((...((((((....((((........)))).(((((.......))))).......)))))).....((....)))).)))))))...... ( -37.00) >DroAna_CAF1 38382 119 + 1 AUAUGAAUGAGCAAAAAGUCGGGCGGAAGUUGUAAGACCCUUUGGCGCAGCGGAUAGCGCGUUGGACUUCUAAUCCAAAGGUCGCGGGUUCGAUCCCCGCAAGGGUUGAAA-ACACAAAC ..........((.....(((.(((....)))....)))((((((((((........))))(((((....))))).))))))..))(..(((((..(((....)))))))).-.)...... ( -35.80) >consensus _________________AGCAGCCGAUUCUCGUAGGACCCUUUAGCGCAUUGGAUAGCGCGUUGGACUUCUAAUCCAAAGGUGGCGGGUUCGAUUCCCGCAAGGGUUGA___________ ...................................(((((((..((((........)))).(((((.......))))).....(((((.......))))))))))))............. (-28.62 = -28.62 + -0.00)

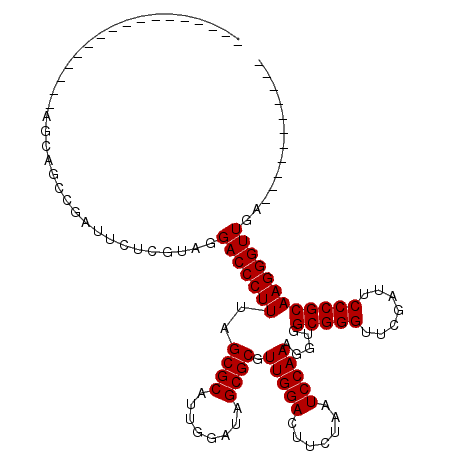
| Location | 1,965,407 – 1,965,499 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.80 |
| Mean single sequence MFE | -31.93 |
| Consensus MFE | -28.30 |
| Energy contribution | -28.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.943428 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1965407 92 - 22407834 -----------UCAACCCUUGCGGGAAUCGAACCCGCCACCUUUGGAUUAGAAGUCCAACGCGCUAUCCAAUGCGCUAAAGGGUCCAACGAGAAUCCGUUGCU----------------- -----------...(((((((((((.......))))).....((((((.....)))))).((((........))))..)))))).(((((......)))))..----------------- ( -32.40) >DroSec_CAF1 7870 92 - 1 -----------UCAACCCUUGCGGGAAUCGAACCCGCCACCUUUGGAUUAGAAGUCCAACGCGCUAUCCAAUGCGCUAAAGGGUCCAACGAGAAUCCGCUGCU----------------- -----------.........(((((..(((.((((.......((((((.....)))))).((((........))))....))))....)))...)))))....----------------- ( -28.50) >DroSim_CAF1 12009 92 - 1 -----------UCAACCCUUGCGGGAAUCGAACCCGCCACCUUUGGAUUAGAAGUCCAACGCGCUAUCCAAUGCGCUAAAGGGUCCAACGAGAAUCCGCUGCU----------------- -----------.........(((((..(((.((((.......((((((.....)))))).((((........))))....))))....)))...)))))....----------------- ( -28.50) >DroEre_CAF1 17042 103 - 1 GCUAAUGGCAAUCAACCCUUGCGGGAAUCGAACCCGCCACCUUUGGAUUAGAAGUCCAACGCGCUAUCCAAUGCGCUAAAGGGUCCUACGAGAUUCGCCGGAU----------------- .....((((((((..((((((((((.......))))).....((((((.....)))))).((((........))))..)))))((....)))))).))))...----------------- ( -33.90) >DroYak_CAF1 14896 103 - 1 UCUAGUGGCAAUCAACCCUUGCGGGAAUCGAACCCGCCACCUUUGGAUUAGAAGUCCAACGCGCUAUCCAAUGCGCUAAAGGGUCCUACGCGAAUCGCCGCAU----------------- ....(((((......((((((((((.......))))).....((((((.....)))))).((((........))))..)))))((......))...)))))..----------------- ( -35.30) >DroAna_CAF1 38382 119 - 1 GUUUGUGU-UUUCAACCCUUGCGGGGAUCGAACCCGCGACCUUUGGAUUAGAAGUCCAACGCGCUAUCCGCUGCGCCAAAGGGUCUUACAACUUCCGCCCGACUUUUUGCUCAUUCAUAU (..((.((-.(((((...(((((((.......)))))))...)))))..(((((((....((((........))))....((((............))))))))))).)).))..).... ( -33.00) >consensus ___________UCAACCCUUGCGGGAAUCGAACCCGCCACCUUUGGAUUAGAAGUCCAACGCGCUAUCCAAUGCGCUAAAGGGUCCAACGAGAAUCCCCUGAU_________________ ..............(((((((((((.......))))).....((((((.....)))))).((((........))))..)))))).................................... (-28.30 = -28.30 + 0.00)
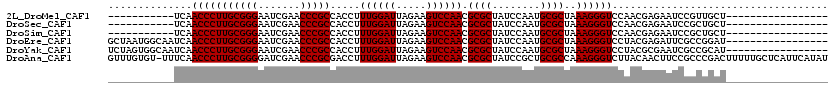
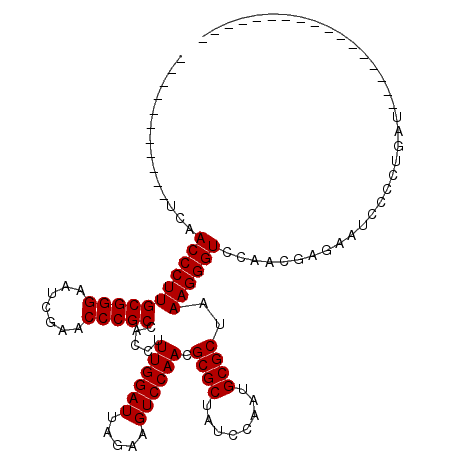
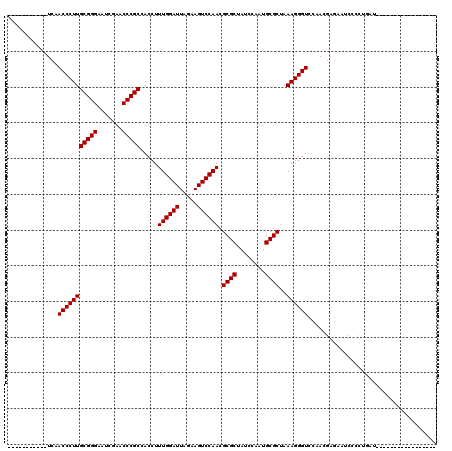
| Location | 1,965,430 – 1,965,527 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.76 |
| Mean single sequence MFE | -26.97 |
| Consensus MFE | -21.10 |
| Energy contribution | -21.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970535 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1965430 97 - 22407834 CU--UUA-GCAU-----GUACA----AAAAAAGGAGUACU-----------UCAACCCUUGCGGGAAUCGAACCCGCCACCUUUGGAUUAGAAGUCCAACGCGCUAUCCAAUGCGCUAAA ..--...-....-----.....----....((((.((...-----------...))))))(((((.......))))).....((((((.....)))))).((((........)))).... ( -24.40) >DroSec_CAF1 7893 91 - 1 CC--UAA------------ACA----AAAAAAGGAGUACU-----------UCAACCCUUGCGGGAAUCGAACCCGCCACCUUUGGAUUAGAAGUCCAACGCGCUAUCCAAUGCGCUAAA ..--...------------...----....((((.((...-----------...))))))(((((.......))))).....((((((.....)))))).((((........)))).... ( -24.40) >DroSim_CAF1 12032 98 - 1 CC--CUAAGCAU-----GUACA----AAAAAAGGAGUACU-----------UCAACCCUUGCGGGAAUCGAACCCGCCACCUUUGGAUUAGAAGUCCAACGCGCUAUCCAAUGCGCUAAA ..--........-----.....----....((((.((...-----------...))))))(((((.......))))).....((((((.....)))))).((((........)))).... ( -24.40) >DroEre_CAF1 17065 103 - 1 AC--C-----------UUUGCC----AAAAAAGUAGCACAGCUAAUGGCAAUCAACCCUUGCGGGAAUCGAACCCGCCACCUUUGGAUUAGAAGUCCAACGCGCUAUCCAAUGCGCUAAA ..--.-----------.(((((----(......((((...)))).)))))).........(((((.......))))).....((((((.....)))))).((((........)))).... ( -29.00) >DroYak_CAF1 14919 118 - 1 GC--CAAAGCAUUUUAUUUGCCAGGCAAAAAAGUAGUACAUCUAGUGGCAAUCAACCCUUGCGGGAAUCGAACCCGCCACCUUUGGAUUAGAAGUCCAACGCGCUAUCCAAUGCGCUAAA ((--(...(((.......)))..))).......(((..(((...(((((...........(((((.......))))).....((((((.....))))))...)))))...)))..))).. ( -31.40) >DroAna_CAF1 38422 104 - 1 ACAUC-----------UUUUAA----AUAUAAGUAACACAGUUUGUGU-UUUCAACCCUUGCGGGGAUCGAACCCGCGACCUUUGGAUUAGAAGUCCAACGCGCUAUCCGCUGCGCCAAA ....(-----------((((((----........(((((.....))))-)(((((...(((((((.......)))))))...))))))))))))......((((........)))).... ( -28.20) >consensus CC__C_A__________UUACA____AAAAAAGGAGUACA___________UCAACCCUUGCGGGAAUCGAACCCGCCACCUUUGGAUUAGAAGUCCAACGCGCUAUCCAAUGCGCUAAA ............................................................(((((.......))))).....((((((.....)))))).((((........)))).... (-21.10 = -21.10 + -0.00)

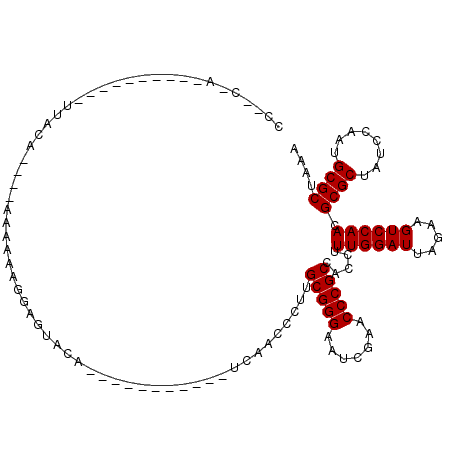
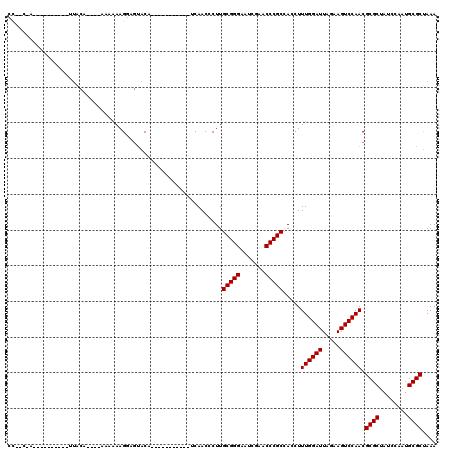
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:42:22 2006