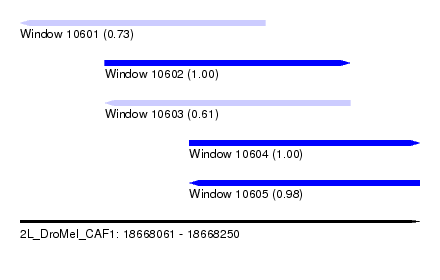
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,668,061 – 18,668,250 |
| Length | 189 |
| Max. P | 0.999707 |

| Location | 18,668,061 – 18,668,177 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 92.41 |
| Mean single sequence MFE | -33.88 |
| Consensus MFE | -26.14 |
| Energy contribution | -26.14 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.726070 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18668061 116 - 22407834 GUAAACAGCUGUAUGGAUCAGAGACUAAUGCUUAACUGUAAGCGCGAGGAUCUGCUGAGGAAGGAGUUAAUGGAACUGAAAACCAAGCUGGAACAGGACAUGACGGUGCAGCGCAG .......(((((((.(..((((..((..((((((....))))))..))..))))(((.......((((.....)))).....((.....))..))).......).))))))).... ( -29.60) >DroSec_CAF1 20738 116 - 1 GUAAACAGCUGUAUGGAUCAGCGACUAAUGCUUAACAGUAAGCGGGAGGAUCUGCUGAGAAAGGAGUUGAUGGAACUGGAAACCAAGCUGAAACAGGACAUGACGGUGCAGCGCAG .......(((((((.(.(((((..(((..((((.......(((((......))))).......))))...)))....(....)...)))))..((.....)).).))))))).... ( -30.44) >DroSim_CAF1 22015 116 - 1 GUAAACAGCUGUAUGGAUCAGCGACUAAUGCUCAACAGUAAGCGGGAGGAUCUGCUGAGAAAGGAGUUGAUGGAACUGGAAACCAAGCUGAAACAGGACAUGACGGUGCAGCGCAG .......(((((((.(.(((((..(((..((((.......(((((......))))).......))))...)))....(....)...)))))..((.....)).).))))))).... ( -32.24) >DroEre_CAF1 20969 116 - 1 GUAAACAGCUGCAUGGAUCAGCGACUAAUGCUUAUCAGCAGCCGCGAGGAGCUGCUGAGAAAGGAGUUGAAGGAACUGGAAACCAAGUUGCAACAGGACAUGACGGUGCAGCGCAG .......(((((((.(.(((.(((((....(((.(((((((((....)..))))))))..))).)))))..(.(((((....)..)))).).........)))).))))))).... ( -40.70) >DroYak_CAF1 20906 116 - 1 GUAAACAGCUGCAUGGAUCAGCGACUAAUGCUUACCAGUAGCCGUGAGGAGCUGCUGAGGAAGGAGUUGAAGGAACUGGAAACCAAGUUGAAACAGGACAUGACGGUGCAGCGCAG .......(((((((.(.(((((..((..(.((((.((((..((....)).)))).)))).)))..)))))...(((((....)..))))..............).))))))).... ( -36.40) >consensus GUAAACAGCUGUAUGGAUCAGCGACUAAUGCUUAACAGUAAGCGCGAGGAUCUGCUGAGAAAGGAGUUGAUGGAACUGGAAACCAAGCUGAAACAGGACAUGACGGUGCAGCGCAG .......(((((((.(.(((.(((((....((((.(((....(....)...))).)))).....)))))...(..(((...............)))..).)))).))))))).... (-26.14 = -26.14 + 0.00)


| Location | 18,668,101 – 18,668,217 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.51 |
| Mean single sequence MFE | -35.07 |
| Consensus MFE | -19.23 |
| Energy contribution | -20.98 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.59 |
| Structure conservation index | 0.55 |
| SVM decision value | 3.03 |
| SVM RNA-class probability | 0.998208 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18668101 116 + 22407834 GUUCCAUUAACUCCUUCCUCAGCAGAUCCUCGCGCUUACAGUUAA----GCAUUAGUCUCUGAUCCAUACAGCUGUUUACCCACUUGAUAUGGUAACGCAUCUGUAUCGGAUCAGUCACU .....................((........))(((((....)))----))...(((..(((((((((((((.((((((((..........))))).))).)))))).)))))))..))) ( -31.20) >DroSec_CAF1 20778 116 + 1 GUUCCAUCAACUCCUUUCUCAGCAGAUCCUCCCGCUUACUGUUAA----GCAUUAGUCGCUGAUCCAUACAGCUGUUUACCCACUUGAUGUGGUAACGCAUCUGUAUCGGAUCAGUCACU .................................(((((....)))----))...(((.((((((((((((((.((((((((.((.....))))))).))).)))))).)))))))).))) ( -34.50) >DroSim_CAF1 22055 116 + 1 GUUCCAUCAACUCCUUUCUCAGCAGAUCCUCCCGCUUACUGUUGA----GCAUUAGUCGCUGAUCCAUACAGCUGUUUACCCACUUGAUGUGGUAACGCAUCUGUAUCGGAUCAGUCACU .................((((((((.............)))))))----)....(((.((((((((((((((.((((((((.((.....))))))).))).)))))).)))))))).))) ( -38.92) >DroEre_CAF1 21009 116 + 1 GUUCCUUCAACUCCUUUCUCAGCAGCUCCUCGCGGCUGCUGAUAA----GCAUUAGUCGCUGAUCCAUGCAGCUGUUUACCCACUUGAUCUGGUAACGCACCUGUAUGGGAUCAGUAGCU .............(((..(((((((((......))))))))).))----)....(((..(((((((((((((.((((((((..........))))).))).)))))).)))))))..))) ( -43.50) >DroWil_CAF1 81793 98 + 1 ----------CUCCCGUUC---CAGAUUAUCCAG-UUUCUGUUGGAUUCUCAUCAUAUACUGCUCGACACCAGUGCGAACCCAUGUGAUGCGAUAGCGCAACUCAAUGGGAG-------- ----------((((((((.---..((..((((((-......))))))..))(((((((...(.(((.((....))))).)..)))))))(((....))).....))))))))-------- ( -28.00) >DroYak_CAF1 20946 116 + 1 GUUCCUUCAACUCCUUCCUCAGCAGCUCCUCACGGCUACUGGUAA----GCAUUAGUCGCUGAUCCAUGCAGCUGUUUACCCACUUAAUCUGGUAACGCACCUGUAUGGGAUCAGAAGCU .............(((.(.(((.((((......)))).)))).))----)....(((..(((((((((((((.((((((((..........))))).))).)))))).)))))))..))) ( -34.30) >consensus GUUCCAUCAACUCCUUUCUCAGCAGAUCCUCCCGCUUACUGUUAA____GCAUUAGUCGCUGAUCCAUACAGCUGUUUACCCACUUGAUGUGGUAACGCAUCUGUAUCGGAUCAGUCACU ......................................................(((..(((((((((((((.((((((((..........))))).))).)))))).)))))))..))) (-19.23 = -20.98 + 1.75)
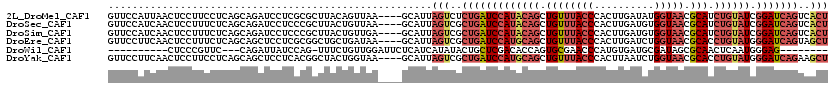
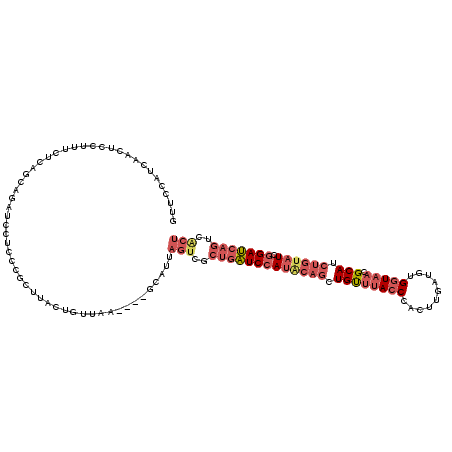

| Location | 18,668,101 – 18,668,217 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.51 |
| Mean single sequence MFE | -35.21 |
| Consensus MFE | -15.23 |
| Energy contribution | -16.85 |
| Covariance contribution | 1.62 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.609443 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18668101 116 - 22407834 AGUGACUGAUCCGAUACAGAUGCGUUACCAUAUCAAGUGGGUAAACAGCUGUAUGGAUCAGAGACUAAUGC----UUAACUGUAAGCGCGAGGAUCUGCUGAGGAAGGAGUUAAUGGAAC ..((((((((((.((((((.((.....((((.....)))).....)).)))))))))))(((..((..(((----(((....))))))..))..)))...........)))))....... ( -32.90) >DroSec_CAF1 20778 116 - 1 AGUGACUGAUCCGAUACAGAUGCGUUACCACAUCAAGUGGGUAAACAGCUGUAUGGAUCAGCGACUAAUGC----UUAACAGUAAGCGGGAGGAUCUGCUGAGAAAGGAGUUGAUGGAAC ..((.(((((((.((((((.((..(((((.(((...)))))))).)).))))))))))))))).(((..((----((.......(((((......))))).......))))...)))... ( -35.44) >DroSim_CAF1 22055 116 - 1 AGUGACUGAUCCGAUACAGAUGCGUUACCACAUCAAGUGGGUAAACAGCUGUAUGGAUCAGCGACUAAUGC----UCAACAGUAAGCGGGAGGAUCUGCUGAGAAAGGAGUUGAUGGAAC ..((.(((((((.((((((.((..(((((.(((...)))))))).)).))))))))))))))).(((..((----((.......(((((......))))).......))))...)))... ( -37.24) >DroEre_CAF1 21009 116 - 1 AGCUACUGAUCCCAUACAGGUGCGUUACCAGAUCAAGUGGGUAAACAGCUGCAUGGAUCAGCGACUAAUGC----UUAUCAGCAGCCGCGAGGAGCUGCUGAGAAAGGAGUUGAAGGAAC .....(((((((.((.(((.((..(((((..........))))).)).))).)))))))))(((((....(----((.(((((((((....)..))))))))..))).)))))....... ( -39.90) >DroWil_CAF1 81793 98 - 1 --------CUCCCAUUGAGUUGCGCUAUCGCAUCACAUGGGUUCGCACUGGUGUCGAGCAGUAUAUGAUGAGAAUCCAACAGAAA-CUGGAUAAUCUG---GAACGGGAG---------- --------(((((.....((((.(...((.(((((.(((.(((((((....)).)))))..))).))))).))..))))).....-(..(.....)..---)...)))))---------- ( -28.90) >DroYak_CAF1 20946 116 - 1 AGCUUCUGAUCCCAUACAGGUGCGUUACCAGAUUAAGUGGGUAAACAGCUGCAUGGAUCAGCGACUAAUGC----UUACCAGUAGCCGUGAGGAGCUGCUGAGGAAGGAGUUGAAGGAAC .....(((((((.((.(((.((..(((((..........))))).)).))).)))))))))(((((..(.(----(((.((((..((....)).)))).)))).)...)))))....... ( -36.90) >consensus AGUGACUGAUCCCAUACAGAUGCGUUACCACAUCAAGUGGGUAAACAGCUGUAUGGAUCAGCGACUAAUGC____UUAACAGUAAGCGGGAGGAUCUGCUGAGAAAGGAGUUGAUGGAAC .....(((((((.((.(((.((..(((((..........))))).)).))).)))))))))(((((..(......(((.(((....(....)...))).)))...)..)))))....... (-15.23 = -16.85 + 1.62)

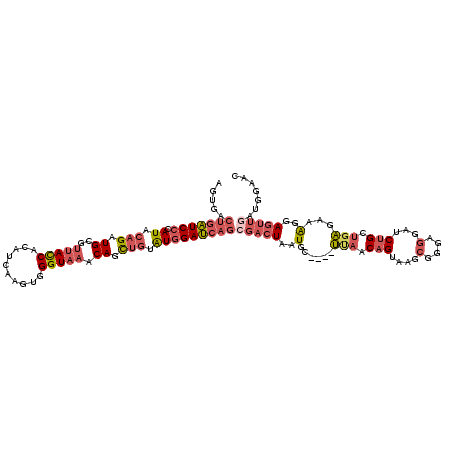

| Location | 18,668,141 – 18,668,250 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 84.11 |
| Mean single sequence MFE | -34.24 |
| Consensus MFE | -29.92 |
| Energy contribution | -29.64 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.08 |
| Mean z-score | -4.13 |
| Structure conservation index | 0.87 |
| SVM decision value | 3.92 |
| SVM RNA-class probability | 0.999707 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18668141 109 + 22407834 GUUAAGCAUUAGUCUCUGAUCCAUACAGCUGUUUACCCACUUGAUAUGGUAACGCAUCUGUAUCGGAUCAGUCACUAUGAAAAUGUACAAAAGAUACAUUAA-----AUUUUUG-- ......(((.(((..(((((((((((((.((((((((..........))))).))).)))))).)))))))..))))))..(((((((....).))))))..-----.......-- ( -30.40) >DroSec_CAF1 20818 107 + 1 GUUAAGCAUUAGUCGCUGAUCCAUACAGCUGUUUACCCACUUGAUGUGGUAACGCAUCUGUAUCGGAUCAGUCACUAAGAAAAUGUACAAAAAAUGUAUC---------UUUUGGU ........(((((.((((((((((((((.((((((((.((.....))))))).))).)))))).)))))))).))))).((((.(((((.....))))).---------))))... ( -36.60) >DroSim_CAF1 22095 107 + 1 GUUGAGCAUUAGUCGCUGAUCCAUACAGCUGUUUACCCACUUGAUGUGGUAACGCAUCUGUAUCGGAUCAGUCACUAAGAAAAUGUACAAAAAAUGUAUC---------UUUUGGU ........(((((.((((((((((((((.((((((((.((.....))))))).))).)))))).)))))))).))))).((((.(((((.....))))).---------))))... ( -36.60) >DroEre_CAF1 21049 110 + 1 GAUAAGCAUUAGUCGCUGAUCCAUGCAGCUGUUUACCCACUUGAUCUGGUAACGCACCUGUAUGGGAUCAGUAGCUAAGAAAUUAUACAAAAGAUAGAUUAC-----A-UUUUUGU .((((...(((((..(((((((((((((.((((((((..........))))).))).)))))).)))))))..)))))....))))(((((((.........-----.-))))))) ( -33.70) >DroYak_CAF1 20986 114 + 1 GGUAAGCAUUAGUCGCUGAUCCAUGCAGCUGUUUACCCACUUAAUCUGGUAACGCACCUGUAUGGGAUCAGAAGCUAAGAAAUUAUAUAAAAUAUACCUUCUAAAAAA-GUUUGG- ..(((((..((((..(((((((((((((.((((((((..........))))).))).)))))).)))))))..))))((((..(((((...)))))..))))......-))))).- ( -33.90) >consensus GUUAAGCAUUAGUCGCUGAUCCAUACAGCUGUUUACCCACUUGAUCUGGUAACGCAUCUGUAUCGGAUCAGUCACUAAGAAAAUGUACAAAAAAUACAUU_______A_UUUUGGU ........(((((..(((((((((((((.((((((((..........))))).))).)))))).)))))))..)))))...................................... (-29.92 = -29.64 + -0.28)


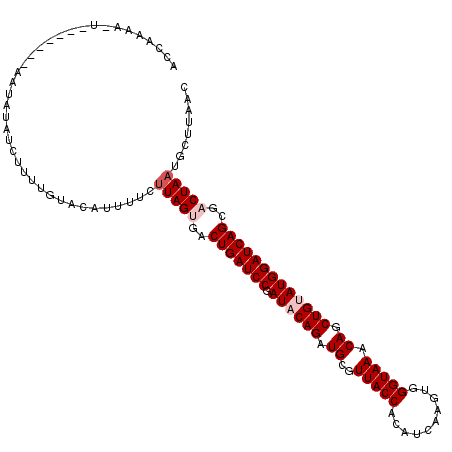
| Location | 18,668,141 – 18,668,250 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 84.11 |
| Mean single sequence MFE | -27.88 |
| Consensus MFE | -23.50 |
| Energy contribution | -24.50 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.983260 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18668141 109 - 22407834 --CAAAAAU-----UUAAUGUAUCUUUUGUACAUUUUCAUAGUGACUGAUCCGAUACAGAUGCGUUACCAUAUCAAGUGGGUAAACAGCUGUAUGGAUCAGAGACUAAUGCUUAAC --.......-----..(((((((.....)))))))....((((..(((((((.((((((.((.....((((.....)))).....)).)))))))))))))..))))......... ( -29.70) >DroSec_CAF1 20818 107 - 1 ACCAAAA---------GAUACAUUUUUUGUACAUUUUCUUAGUGACUGAUCCGAUACAGAUGCGUUACCACAUCAAGUGGGUAAACAGCUGUAUGGAUCAGCGACUAAUGCUUAAC ...((((---------..((((.....))))..)))).(((((..(((((((.((((((.((..(((((.(((...)))))))).)).)))))))))))))..)))))........ ( -31.50) >DroSim_CAF1 22095 107 - 1 ACCAAAA---------GAUACAUUUUUUGUACAUUUUCUUAGUGACUGAUCCGAUACAGAUGCGUUACCACAUCAAGUGGGUAAACAGCUGUAUGGAUCAGCGACUAAUGCUCAAC ...((((---------..((((.....))))..)))).(((((..(((((((.((((((.((..(((((.(((...)))))))).)).)))))))))))))..)))))........ ( -31.50) >DroEre_CAF1 21049 110 - 1 ACAAAAA-U-----GUAAUCUAUCUUUUGUAUAAUUUCUUAGCUACUGAUCCCAUACAGGUGCGUUACCAGAUCAAGUGGGUAAACAGCUGCAUGGAUCAGCGACUAAUGCUUAUC (((((((-(-----(.....))).))))))........((((...(((((((.((.(((.((..(((((..........))))).)).))).)))))))))...))))........ ( -22.80) >DroYak_CAF1 20986 114 - 1 -CCAAAC-UUUUUUAGAAGGUAUAUUUUAUAUAAUUUCUUAGCUUCUGAUCCCAUACAGGUGCGUUACCAGAUUAAGUGGGUAAACAGCUGCAUGGAUCAGCGACUAAUGCUUACC -......-.......((((.(((((...))))).))))((((...(((((((.((.(((.((..(((((..........))))).)).))).)))))))))...))))........ ( -23.90) >consensus ACCAAAA_U_______AAUAUAUCUUUUGUACAUUUUCUUAGUGACUGAUCCGAUACAGAUGCGUUACCACAUCAAGUGGGUAAACAGCUGUAUGGAUCAGCGACUAAUGCUUAAC ......................................(((((..(((((((.((((((.((..(((((..........))))).)).)))))))))))))..)))))........ (-23.50 = -24.50 + 1.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:16:25 2006