| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,657,119 – 18,657,209 |
| Length | 90 |
| Max. P | 0.956860 |

| Location | 18,657,119 – 18,657,209 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 75.70 |
| Mean single sequence MFE | -27.92 |
| Consensus MFE | -24.49 |
| Energy contribution | -23.47 |
| Covariance contribution | -1.02 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956860 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
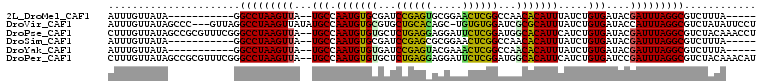
>2L_DroMel_CAF1 18657119 90 + 22407834 AUUUGUUAUA-----------GGCCUAAGUUA--UGCCAAUGUGCGAUCCGAGUGCGGAACUCGGCCAACACAUUUAUCUGUGAUACGAUUUAGGCGUCUUUA----- ..........-----------.(((((((((.--....((((((....((((((.....))))))....))))))((((...)))).))))))))).......----- ( -25.40) >DroVir_CAF1 57020 104 + 1 AUUUGUUAUAGCCC---GUUAGGCCUAAGUUAUAUGCCAAUGUGCGUGCUGCACAGC-UGUGUGGAUCGCGCAUUUAUCUGUGAUACCAUUUAGGCGUCUAUAUUCCU ......(((((...---...(.((((((((((((....((((((((..(..(((...-.)))..)..))))))))....)))))).....)))))).))))))..... ( -28.40) >DroPse_CAF1 64333 106 + 1 CUUUGUUAUAGCCGCGUUUCGGGCCUAAGUUA--UGCCAAUGUGUGCUCUGAGGAGGAUUCUCGGAUGGCACAUUCAUCUGUGAUACGAUUUAGGCGUCUACAAACCU .(((((....(((.(((.((((((........--.)))(((((((..((((((((...))))))))..)))))))......))).))).....)))....)))))... ( -30.90) >DroSim_CAF1 10256 90 + 1 AUUUGUUAUA-----------GGCCUAAGUUA--UGCCAAUGUGCGAUCCGAGCGCGGAACUCGGCCAACACAUUUAUCUGUGAUACGAUUUAGGCGUCUUUA----- ..........-----------.(((((((((.--.(((..(((((.......)))))(....))))...((((......))))....))))))))).......----- ( -23.50) >DroYak_CAF1 9789 90 + 1 AUUUGUUAUA-----------GGCCUAAGUUA--UGCCAAUGUGUGAUCCGAGUACGAAACUCGGCCAACACAUUUAUCUGUGAUACGAUUUAGGCGUCUUUA----- ..........-----------.(((((((((.--....(((((((...((((((.....))))))...)))))))((((...)))).))))))))).......----- ( -27.40) >DroPer_CAF1 52533 106 + 1 CUUUGUUAUAGCCGCGUUUCGGGCCUAAGUUA--UGCCAAUGUGUGCUCUGAGGAGGAUUCUCGGAUGGCACAUUCAUCUGUGAUCCGAUUUAGGCGUCUACAAACAU .(((((....(((.....((((..(..((..(--((..(((((((..((((((((...))))))))..))))))))))))..)..))))....)))....)))))... ( -31.90) >consensus AUUUGUUAUA___________GGCCUAAGUUA__UGCCAAUGUGCGAUCCGAGCACGAAACUCGGACAACACAUUUAUCUGUGAUACGAUUUAGGCGUCUAUA_____ ......................(((((((((...(((.(((((((...((((((.....))))))...))))))).....)))....)))))))))............ (-24.49 = -23.47 + -1.02)
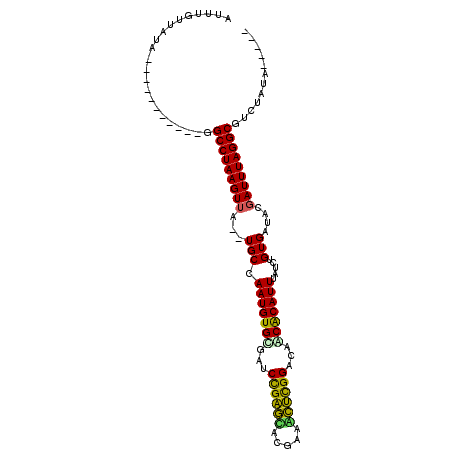
| Location | 18,657,119 – 18,657,209 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 75.70 |
| Mean single sequence MFE | -25.07 |
| Consensus MFE | -15.33 |
| Energy contribution | -15.58 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916634 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18657119 90 - 22407834 -----UAAAGACGCCUAAAUCGUAUCACAGAUAAAUGUGUUGGCCGAGUUCCGCACUCGGAUCGCACAUUGGCA--UAACUUAGGCC-----------UAUAACAAAU -----.......(((((((((........))).(((((((...((((((.....))))))...)))))))....--....)))))).-----------.......... ( -25.20) >DroVir_CAF1 57020 104 - 1 AGGAAUAUAGACGCCUAAAUGGUAUCACAGAUAAAUGCGCGAUCCACACA-GCUGUGCAGCACGCACAUUGGCAUAUAACUUAGGCCUAAC---GGGCUAUAACAAAU .........((..((.....))..))........((((((..........-))(((((.....)))))...))))......(((.((....---)).)))........ ( -22.90) >DroPse_CAF1 64333 106 - 1 AGGUUUGUAGACGCCUAAAUCGUAUCACAGAUGAAUGUGCCAUCCGAGAAUCCUCCUCAGAGCACACAUUGGCA--UAACUUAGGCCCGAAACGCGGCUAUAACAAAG ....((((((..(((((((((........)))...(((((((((.(((.......))).))........)))))--))..))))))(((.....)))))))))..... ( -24.50) >DroSim_CAF1 10256 90 - 1 -----UAAAGACGCCUAAAUCGUAUCACAGAUAAAUGUGUUGGCCGAGUUCCGCGCUCGGAUCGCACAUUGGCA--UAACUUAGGCC-----------UAUAACAAAU -----.......(((((((((........))).(((((((...((((((.....))))))...)))))))....--....)))))).-----------.......... ( -25.50) >DroYak_CAF1 9789 90 - 1 -----UAAAGACGCCUAAAUCGUAUCACAGAUAAAUGUGUUGGCCGAGUUUCGUACUCGGAUCACACAUUGGCA--UAACUUAGGCC-----------UAUAACAAAU -----.......(((((((((........))).(((((((.((((((((.....)))))).)))))))))....--....)))))).-----------.......... ( -25.70) >DroPer_CAF1 52533 106 - 1 AUGUUUGUAGACGCCUAAAUCGGAUCACAGAUGAAUGUGCCAUCCGAGAAUCCUCCUCAGAGCACACAUUGGCA--UAACUUAGGCCCGAAACGCGGCUAUAACAAAG .((((.((((..((((((.((((((((((......))))..))))))..............((........)).--....))))))(((.....)))))))))))... ( -26.60) >consensus _____UAAAGACGCCUAAAUCGUAUCACAGAUAAAUGUGCUAGCCGAGUAUCCUACUCAGAUCACACAUUGGCA__UAACUUAGGCC___________UAUAACAAAU ............(((((((((........))).(((((((...(.((((.....)))).)...)))))))..........))))))...................... (-15.33 = -15.58 + 0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:16:17 2006