
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,642,825 – 18,643,005 |
| Length | 180 |
| Max. P | 0.963205 |

| Location | 18,642,825 – 18,642,945 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.98 |
| Mean single sequence MFE | -31.74 |
| Consensus MFE | -23.03 |
| Energy contribution | -23.19 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.842526 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18642825 120 + 22407834 AAAGGAAACUCACGGAUGGCGAUUCCAUUAUGUAUGUAUGUACAUCCCAGAUUCCGUAUGCACUCAUUCCAAGUGACAAACUUGGCAAACUUGGGCAAUUUCCGCUCCACGGAAAUGUGC ..((....)).((((((((.((........(((((....))))))))))...)))))..((((....(((((((..((....))....)))))))..(((((((.....))))))))))) ( -31.00) >DroSec_CAF1 115574 111 + 1 AAAGGAAACUCAUGGAUGGCGAUUCUAUUAUGUAUGUACGCACAUCCCAGAUUCCGUAUGCACUCAUUCCAAGUGACAAA---------CUUGGGCAAUUUCCGCUCCACGGAAAUGUGC ...((((.((...((((((((((.(......)...)).))).))))).)).))))....((((....(((((((.....)---------))))))..(((((((.....))))))))))) ( -34.30) >DroSim_CAF1 117201 111 + 1 AAAGGAAACUCAUGGAUGGCGAUUCCAUUAUGUAUGUACGCACAUCCCAGAUUCCGUAUGCACUCAUUCCAAGUGACAAA---------CUUGGGCAAUUUCCGCUCCACGGAAAUGUGC ...((((.((...((((((((....(((.....)))..))).))))).)).))))....((((....(((((((.....)---------))))))..(((((((.....))))))))))) ( -35.10) >DroEre_CAF1 114533 103 + 1 AAAGGAAACUCAUGGAUGGCGAUUCCAUUAUGU--------ACAUCCCCGAUUCCGUAUGUACUCAUUCCAAGUGACAAA---------CUUGGGCAAUUUCCGCUCCACGGAAAUGUGC ..((....)).(((((.......)))))...((--------((((...((....)).)))))).....((((((.....)---------)))))((((((((((.....))))))).))) ( -27.70) >DroYak_CAF1 115967 103 + 1 AGAGGAAACUCAUAGAUGGCGAUUCCAUUAUGU--------ACAUCCCAGAUUCCGUAUGUACUCAUUCCAAGUGACAAA---------CUUGGGCAAUUUCCGCUCCACGGAAAUGUGC .(((....)))...(((((.....)))))..((--------((((.(........).)))))).....((((((.....)---------)))))((((((((((.....))))))).))) ( -30.60) >consensus AAAGGAAACUCAUGGAUGGCGAUUCCAUUAUGUAUGUA_G_ACAUCCCAGAUUCCGUAUGCACUCAUUCCAAGUGACAAA_________CUUGGGCAAUUUCCGCUCCACGGAAAUGUGC ...((((.((...(((((........................))))).)).))))....((((....((((((................))))))..(((((((.....))))))))))) (-23.03 = -23.19 + 0.16)

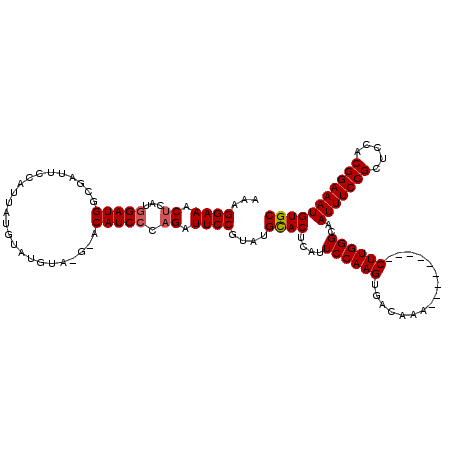
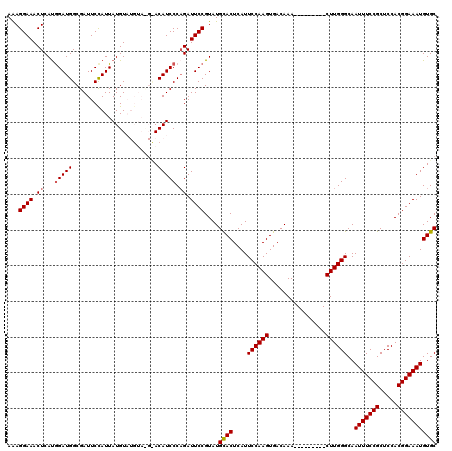
| Location | 18,642,905 – 18,643,005 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 81.90 |
| Mean single sequence MFE | -26.14 |
| Consensus MFE | -21.76 |
| Energy contribution | -21.80 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.804552 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18642905 100 + 22407834 CUUGGCAAACUUGGGCAAUUUCCGCUCCACGGAAAUGUGCACAAUUUAAUUUUGGCCUGCGGUGGCAGGGGUUUGCUCAUUUUUUCCCCC--------GUUUUUCUUU ...((((((((((.((((((((((.....))))))).))).))............(((((....))))))))))))).............--------.......... ( -28.60) >DroSec_CAF1 115654 93 + 1 ---------CUUGGGCAAUUUCCGCUCCACGGAAAUGUGCACAAUUUAAUUUUGGCCUGCGGUAGCAGGGGUUUGCUCACUUCCUCCCUGC------UUUUUUUUUUC ---------...((((.(((((((.....))))))).....(((.......))))))).....((((((((.............)))))))------).......... ( -24.42) >DroSim_CAF1 117281 99 + 1 ---------CUUGGGCAAUUUCCGCUCCACGGAAAUGUGCACAAUUUAAUUUUGGCCUGCGGUAGCAGGGGUUUGCUCACUUACCCCCUCCCCCCAGCUAUUUUUUUC ---------.(((.((((((((((.....))))))).))).)))........((((.((.((..(..(((((..(....)..)))))..)..))))))))........ ( -28.10) >DroEre_CAF1 114605 84 + 1 ---------CUUGGGCAAUUUCCGCUCCACGGAAAUGUGCACAAUUUAAUUUUGGCCUGCGGUGGCAGGGGUUUGCUCAUUUUU-CCCCG---------U-----UUU ---------..(((((((...((.((((((.(....(.((.(((.......))))))..).)))).)).)).))))))).....-.....---------.-----... ( -24.50) >DroYak_CAF1 116039 85 + 1 ---------CUUGGGCAAUUUCCGCUCCACGGAAAUGUGCACAAUUUAAUUUUGGACUGCGGUGGCAGGGGCUUGCUCAUUUUUGCCCCG---------U-----UUU ---------.(((.((((((((((.....))))))).))).)))..............((((.((((((((....)))....))))))))---------)-----... ( -25.10) >consensus _________CUUGGGCAAUUUCCGCUCCACGGAAAUGUGCACAAUUUAAUUUUGGCCUGCGGUGGCAGGGGUUUGCUCAUUUUUUCCCCG_________UUUUU_UUU ...........(((((((((((((.....)))))))..((.(((.......))).(((((....))))).)).))))))............................. (-21.76 = -21.80 + 0.04)

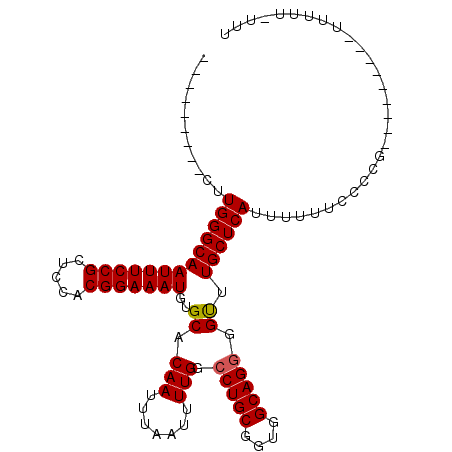
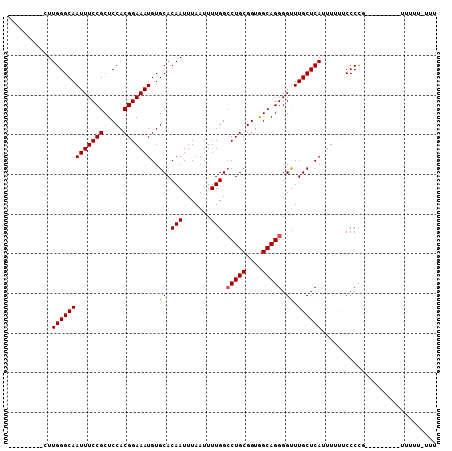
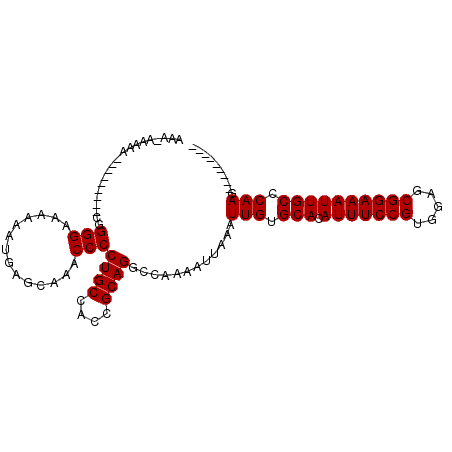
| Location | 18,642,905 – 18,643,005 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 81.90 |
| Mean single sequence MFE | -26.38 |
| Consensus MFE | -19.88 |
| Energy contribution | -19.88 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963205 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18642905 100 - 22407834 AAAGAAAAAC--------GGGGGAAAAAAUGAGCAAACCCCUGCCACCGCAGGCCAAAAUUAAAUUGUGCACAUUUCCGUGGAGCGGAAAUUGCCCAAGUUUGCCAAG ..........--------...........((.((((((.(((((....)))))...........(((.(((.(((((((.....)))))))))).))))))))))).. ( -28.30) >DroSec_CAF1 115654 93 - 1 GAAAAAAAAAA------GCAGGGAGGAAGUGAGCAAACCCCUGCUACCGCAGGCCAAAAUUAAAUUGUGCACAUUUCCGUGGAGCGGAAAUUGCCCAAG--------- ..........(------((((((.(............))))))))...................(((.(((.(((((((.....)))))))))).))).--------- ( -25.30) >DroSim_CAF1 117281 99 - 1 GAAAAAAAUAGCUGGGGGGAGGGGGUAAGUGAGCAAACCCCUGCUACCGCAGGCCAAAAUUAAAUUGUGCACAUUUCCGUGGAGCGGAAAUUGCCCAAG--------- ...........(((.((...((((((..(....)..))))))....)).)))............(((.(((.(((((((.....)))))))))).))).--------- ( -31.60) >DroEre_CAF1 114605 84 - 1 AAA-----A---------CGGGG-AAAAAUGAGCAAACCCCUGCCACCGCAGGCCAAAAUUAAAUUGUGCACAUUUCCGUGGAGCGGAAAUUGCCCAAG--------- ...-----.---------.((((-.............)))).(((......)))..........(((.(((.(((((((.....)))))))))).))).--------- ( -22.62) >DroYak_CAF1 116039 85 - 1 AAA-----A---------CGGGGCAAAAAUGAGCAAGCCCCUGCCACCGCAGUCCAAAAUUAAAUUGUGCACAUUUCCGUGGAGCGGAAAUUGCCCAAG--------- ...-----.---------.(((((............)))))(((....))).............(((.(((.(((((((.....)))))))))).))).--------- ( -24.10) >consensus AAA_AAAAA_________CGGGGAAAAAAUGAGCAAACCCCUGCCACCGCAGGCCAAAAUUAAAUUGUGCACAUUUCCGUGGAGCGGAAAUUGCCCAAG_________ ....................(((..............)))((((....))))............(((.(((.(((((((.....)))))))))).))).......... (-19.88 = -19.88 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:16:14 2006