| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,635,699 – 18,635,818 |
| Length | 119 |
| Max. P | 0.738727 |

| Location | 18,635,699 – 18,635,818 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.49 |
| Mean single sequence MFE | -30.66 |
| Consensus MFE | -18.00 |
| Energy contribution | -18.22 |
| Covariance contribution | 0.21 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.738727 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
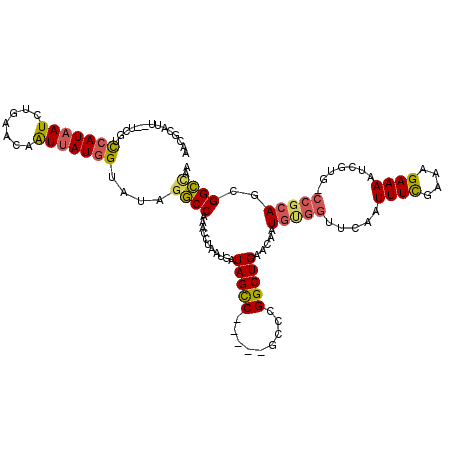
>2L_DroMel_CAF1 18635699 119 - 22407834 AACGCAUUUUUCGUCCAUAAUCUGAACAGUAAUGGUAUAGGCCAAAUCUAAUGAUAGCCUGAUAGCCCGGCUGAACAAUGGGGUUCAAUUUCGAAAGAAAAUCGUG-CCGCAGCGGCCAA ...((.........((((...((....))..))))..(((((...(((....))).)))))...))..(((((.....((.(((....((((....)))).....)-)).)).))))).. ( -30.80) >DroSec_CAF1 108574 117 - 1 AACGCAUU--UCGUUCAUAAUCUGAACAAUUAUGGUAUAGGCCAAACCUAAUGAUAGCCUGAUAGCCCGGCUGAACAAUGUGGUUCAAUUUUGAAAGAAAAUCGUG-CCGCAGCGGCCAA .((.(((.--..(((((.....)))))....)))))...((((...........(((((.(.....).))))).....((((((.(.(((((.....))))).).)-)))))..)))).. ( -29.30) >DroSim_CAF1 110147 117 - 1 AACGCAUU--UCGUUCAUAAUCUGAACAAUUAUGGUAUAGGCCAAACCUAAUGAUAGCCUGAUAGCCCGGCUGAACAAUGUGGUUCAAUUUCGAAAGAAAAUCGUG-CCGGAGCGGCCAA ........--..(((((.....))))).....((((.(((((..............)))))...((((((((((((......))))).((((....)))).....)-)))).)).)))). ( -35.54) >DroEre_CAF1 107490 112 - 1 AACGCAUU--UCGCCCAUAAUCUGAACAAUUAUGGUAUAGGCCAAACCUAAUGAUAGUC-----GCCCGGCUGAACAAUGUGGUUCGAUUUCGAAAGAAAAUCGUG-CCGCAGCGGCAAA ..(((...--(((.(((((((.......)))))))..((((.....)))).)))(((((-----....))))).....((((((.(((((((....).)))))).)-))))))))..... ( -33.40) >DroYak_CAF1 108909 112 - 1 AACGCAUU--UCGUCCAUAAUCUAAACAAUUAUGGUAUAGGCCAAACAUAAUGAUAGUC-----GCCCGGCUGAACAAUGUGGUUCAAUUUCGAAAGAAAAUCGUG-CCGCAGCGGCCAA ........--....(((((((.......)))))))....((((...........(((((-----....))))).....((((((....((((....)))).....)-)))))..)))).. ( -30.80) >DroAna_CAF1 98717 93 - 1 CAGUAAUU--UGGGCAAUAAUCUCAACAAUUAUGGCCCAGACCCGCCCAAAU----------------------ACAAUGCGAUCCAGUUUCGU-GGAAAAUCGCGGCAGCAGUGGUU-- ......((--(((((.(((((.......))))).))))))).....(((...----------------------....(((((((((......)-)))...))))).......)))..-- ( -24.14) >consensus AACGCAUU__UCGUCCAUAAUCUGAACAAUUAUGGUAUAGGCCAAACCUAAUGAUAGCC_____GCCCGGCUGAACAAUGUGGUUCAAUUUCGAAAGAAAAUCGUG_CCGCAGCGGCCAA ..............(((((((.......)))))))....((((...........(((((.........))))).....(((((.....((((....)))).......)))))..)))).. (-18.00 = -18.22 + 0.21)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:16:04 2006