| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,634,615 – 18,634,716 |
| Length | 101 |
| Max. P | 0.573080 |

| Location | 18,634,615 – 18,634,716 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.17 |
| Mean single sequence MFE | -24.75 |
| Consensus MFE | -19.15 |
| Energy contribution | -20.24 |
| Covariance contribution | 1.09 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.573080 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
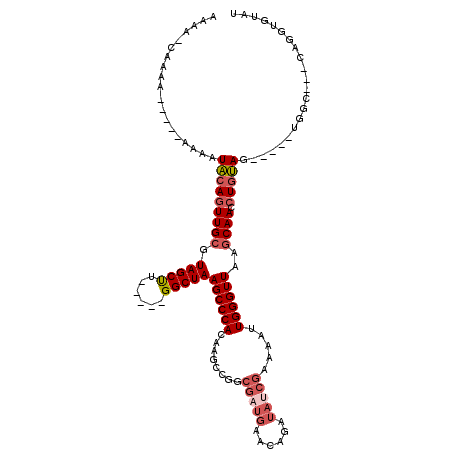
>2L_DroMel_CAF1 18634615 101 + 22407834 AAAA-CAAAA-----AAAAUACAGUUGCGUAGCUU-----GGCUAAGCCCACAAGGCGGCGAUGAACAGAUAUCGAAAAUUGGGUUAAGCAACCUGUAG-----UGGC---CAGGUGUAU ...(-((...-----....(((((((((.(((((.-----.(....(((.....)))..(((((......)))))....)..))))).)))).))))).-----....---....))).. ( -24.69) >DroSec_CAF1 107477 109 + 1 AAAA-CAAAAAAAAUAAAAUACAGUUGCGUAGCUU-----GGCUAAGCCCACAAGGCGGCGAUGAACAGAUAUCGAAAAUUGGGUUAAGCAACCUGUAG-----UGGCCGCCAGGUGUAU ....-.............(((((.((((.((((..-----.)))).))......((((((((((......)))).......(((((....)))))....-----..)))))))).))))) ( -26.90) >DroSim_CAF1 109054 107 + 1 AAAA-CAAAACAA--AAAAUACAAUUGCGUAGCUU-----GGCUAAGCCCACAAGGCGGCGAUGAACAGAUAUCGAAAAUUGGGUUAAGCAACCUGUAG-----UGGCCGCCAGGUGUAU ....-........--...........((.((((..-----.)))).)).(((..((((((((((......)))).......(((((....)))))....-----..))))))..)))... ( -26.50) >DroEre_CAF1 106372 94 + 1 AA-------------AAAAUACAGUUGCGUAGCUU-----GGCUAAGCCCACAAGCCAGCGAUGAACAGAUACCGAAAAUUGGGUUAAGCAACCUGUAG-----UGGC---CAGUUGUAU ..-------------....(((((((((...(((.-----((((.........)))))))............((((...)))).....)))).))))).-----....---......... ( -20.60) >DroYak_CAF1 107779 96 + 1 AAAA-----------AAAAUACAGUUGCGUAGCUU-----GGCUAAGCCCACAAGCCAGCGAUGAACAGAUAUCGAAAAUUGGGUUAAGCAACCUGUAG-----UGGC---CAGGUGUAU ....-----------...(((((.(((.((.(((.-----.(((.((((((........(((((......))))).....)))))).))).......))-----).))---))).))))) ( -23.72) >DroAna_CAF1 97831 105 + 1 AAAAACGCUAAAA--AAAAUGAAGUUGCGUAGCCGGAGCGGGCUAAGCCCACAAGCC----------AGAUAUCGAAAAUUGGGUUAAACAACCUGCACACACCUGGC---CAGGUGUAU ....((((.....--...........)))).((.((.(.((((...)))).).((((----------.(((.......))).))))......)).))..((((((...---.)))))).. ( -26.09) >consensus AAAA_CAAAA_____AAAAUACAGUUGCGUAGCUU_____GGCUAAGCCCACAAGCCGGCGAUGAACAGAUAUCGAAAAUUGGGUUAAGCAACCUGUAG_____UGGC___CAGGUGUAU ...................(((((((((.(((((......)))))((((((........(((((......))))).....))))))..)))).)))))...................... (-19.15 = -20.24 + 1.09)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:16:02 2006