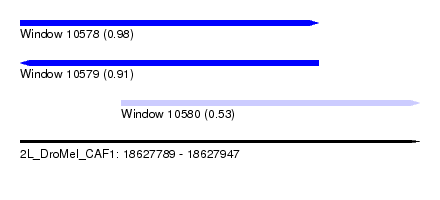
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,627,789 – 18,627,947 |
| Length | 158 |
| Max. P | 0.980612 |

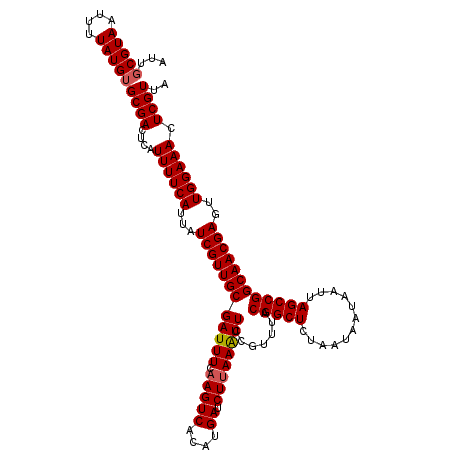
| Location | 18,627,789 – 18,627,907 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 96.95 |
| Mean single sequence MFE | -27.50 |
| Consensus MFE | -25.88 |
| Energy contribution | -26.04 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.980612 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
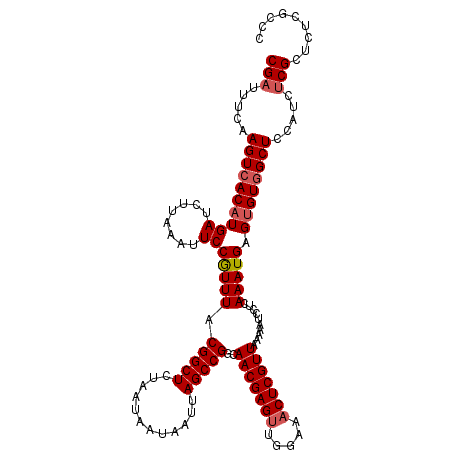
>2L_DroMel_CAF1 18627789 118 + 22407834 AUUGCGUAAUUUUAUGUGCGACUCAUUUUCAUUAUCGUUGCGAUUUCAAGUCACAUGAUCUUAAAUUCCAUUUACGGCUCUAAUAAUAAUUAGCCGGCAACGAGUUGGAAACUCGUUA ...(((((....)))))((((....((((((...((((((((((((.(((((....)).)))))))).......(((((............))))))))))))..)))))).)))).. ( -27.30) >DroSec_CAF1 105283 118 + 1 AAUGCGUAAUUUUAUGUGCGACUCAUUUUCAUUAUCGUUGCGAUUUCAAGUCACAUGAUCUUAAAUUCCGUUUACGGCUCUAAUAAUAAUUAGCCGGCAACGAGUUGGAAACUCGUUA ...(((((....)))))((((....((((((...((((((((((((.(((((....)).)))))))).......(((((............))))))))))))..)))))).)))).. ( -27.30) >DroSim_CAF1 106894 118 + 1 AUUGCGUAAUUUUAUGUGCGACUGAUUUUCAUUAUCGUUGCGAUUUCAAGUCACAUGAUCUUAAAUUCCGUUUACGGCUCUAAUAAUAAUUAGCCGGCAACGAGUUGGAAACUCGUUA ...((.(((..(((((((((((.(((.......))))))))(((.....)))))))))..)))...........(((((............)))))))(((((((.....))))))). ( -28.60) >DroEre_CAF1 104273 118 + 1 AUUUCGUAAUUUUAUGUGCGACUCAUUUUCAUUAUCGUUGCGAUUUCAAGUCACAUGAUCUUAAGUUCCGUUUACGGCUCUAAUAAAAAUUAGCCGGCAACGAGUUGGAAACUCGUUA ...(((((........)))))....((((((...((((((((((((.(((((....)).)))))))).......(((((............))))))))))))..))))))....... ( -26.70) >DroYak_CAF1 105681 118 + 1 AUUGCGUAAUUUUAUGUGCGACUCAUUUUCAUUAUCGUUGCGAUUUCCAGUCACAUGAUCUUAAGUUCCGUUUACGGCUCUAAUAAAAAUUAGCCGGCAACGAGAUGGAAACUCGUUA ...(((((....)))))((((....((((((((.((((((((((.....)))......................(((((............)))))))))))))))))))).)))).. ( -27.60) >consensus AUUGCGUAAUUUUAUGUGCGACUCAUUUUCAUUAUCGUUGCGAUUUCAAGUCACAUGAUCUUAAAUUCCGUUUACGGCUCUAAUAAUAAUUAGCCGGCAACGAGUUGGAAACUCGUUA ...(((((....)))))((((....((((((...((((((((((((.(((((....)).)))))))).......(((((............))))))))))))..)))))).)))).. (-25.88 = -26.04 + 0.16)

| Location | 18,627,789 – 18,627,907 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 96.95 |
| Mean single sequence MFE | -26.14 |
| Consensus MFE | -23.24 |
| Energy contribution | -23.28 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.909068 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18627789 118 - 22407834 UAACGAGUUUCCAACUCGUUGCCGGCUAAUUAUUAUUAGAGCCGUAAAUGGAAUUUAAGAUCAUGUGACUUGAAAUCGCAACGAUAAUGAAAAUGAGUCGCACAUAAAAUUACGCAAU .(((((((.....)))))))((.((((............))))((((.((((.((((((.((....)))))))).)).)).((((.((....))..)))).........))))))... ( -25.10) >DroSec_CAF1 105283 118 - 1 UAACGAGUUUCCAACUCGUUGCCGGCUAAUUAUUAUUAGAGCCGUAAACGGAAUUUAAGAUCAUGUGACUUGAAAUCGCAACGAUAAUGAAAAUGAGUCGCACAUAAAAUUACGCAUU ((((((((.....))))))))...(((((((....(((((.(((....)))..))))).....((((((((...((((...)))).((....)))))))))).....))))).))... ( -25.30) >DroSim_CAF1 106894 118 - 1 UAACGAGUUUCCAACUCGUUGCCGGCUAAUUAUUAUUAGAGCCGUAAACGGAAUUUAAGAUCAUGUGACUUGAAAUCGCAACGAUAAUGAAAAUCAGUCGCACAUAAAAUUACGCAAU ....((.((((....((((((((((((............)))))......((.((((((.((....)))))))).)))))))))....)))).))....((............))... ( -25.60) >DroEre_CAF1 104273 118 - 1 UAACGAGUUUCCAACUCGUUGCCGGCUAAUUUUUAUUAGAGCCGUAAACGGAACUUAAGAUCAUGUGACUUGAAAUCGCAACGAUAAUGAAAAUGAGUCGCACAUAAAAUUACGAAAU ((((((((.....)))))))).(((((.((((((((((..((((....)))............(((((.......))))).)..)))))))))).))))).................. ( -27.80) >DroYak_CAF1 105681 118 - 1 UAACGAGUUUCCAUCUCGUUGCCGGCUAAUUUUUAUUAGAGCCGUAAACGGAACUUAAGAUCAUGUGACUGGAAAUCGCAACGAUAAUGAAAAUGAGUCGCACAUAAAAUUACGCAAU (((((((.......))))))).(((((.((((((((((..((((....)))............(((((.(....)))))).)..)))))))))).))))).................. ( -26.90) >consensus UAACGAGUUUCCAACUCGUUGCCGGCUAAUUAUUAUUAGAGCCGUAAACGGAAUUUAAGAUCAUGUGACUUGAAAUCGCAACGAUAAUGAAAAUGAGUCGCACAUAAAAUUACGCAAU ((((((((.....)))))))).(((((.(((.((((((..((((....)))............(((((.......))))).)..)))))).))).))))).................. (-23.24 = -23.28 + 0.04)

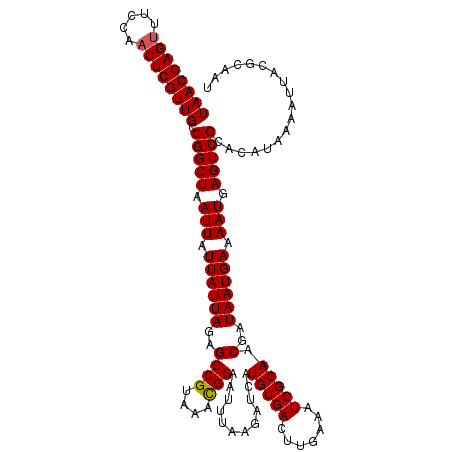

| Location | 18,627,829 – 18,627,947 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 94.92 |
| Mean single sequence MFE | -27.30 |
| Consensus MFE | -22.84 |
| Energy contribution | -23.48 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.534746 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18627829 118 + 22407834 CGAUUUCAAGUCACAUGAUCUUAAAUUCCAUUUACGGCUCUAAUAAUAAUUAGCCGGCAACGAGUUGGAAACUCGUUAAAAAUCCUCAAAUGAGUGUGGCUCCAUCCCGCUCUCGCCC .(((((.(((((....)).))))))))........((((............))))((((((((((.....)))))))..............(((.((((.......)))).)))))). ( -26.80) >DroSec_CAF1 105323 118 + 1 CGAUUUCAAGUCACAUGAUCUUAAAUUCCGUUUACGGCUCUAAUAAUAAUUAGCCGGCAACGAGUUGGAAACUCGUUAAAAAUCCUCAAAUGAGUGUGGCUGCAUCUCGCUCCCGCCC (((.....((((((((((........))(((((.(((((............)))))..(((((((.....)))))))..........))))).)))))))).....)))......... ( -26.50) >DroSim_CAF1 106934 118 + 1 CGAUUUCAAGUCACAUGAUCUUAAAUUCCGUUUACGGCUCUAAUAAUAAUUAGCCGGCAACGAGUUGGAAACUCGUUAAAAAUCCUCAAAUGAGUGUGGCUGCAUCUCGCUCCCGCCC (((.....((((((((((........))(((((.(((((............)))))..(((((((.....)))))))..........))))).)))))))).....)))......... ( -26.50) >DroEre_CAF1 104313 118 + 1 CGAUUUCAAGUCACAUGAUCUUAAGUUCCGUUUACGGCUCUAAUAAAAAUUAGCCGGCAACGAGUUGGAAACUCGUUAAAAAUCCUCAAAUGAGCGUGGCUUCAUCUCGCUCUUUCCC (((..(.(((((((..((........)).((((((((((............)))))..(((((((.....))))))).............)))))))))))).)..)))......... ( -26.30) >DroYak_CAF1 105721 118 + 1 CGAUUUCCAGUCACAUGAUCUUAAGUUCCGUUUACGGCUCUAAUAAAAAUUAGCCGGCAACGAGAUGGAAACUCGUUAAAAAUCCUCAAAUGAGUGUCGCUCCAUCUCGCUCUUGCCC .(((.....)))...........((..(((....)))..))..............((((((((((((((.(((((((...........))))))).....)))))))))...))))). ( -30.40) >consensus CGAUUUCAAGUCACAUGAUCUUAAAUUCCGUUUACGGCUCUAAUAAUAAUUAGCCGGCAACGAGUUGGAAACUCGUUAAAAAUCCUCAAAUGAGUGUGGCUCCAUCUCGCUCUCGCCC (((.....((((((((((........))(((((.(((((............)))))..(((((((.....)))))))..........))))).)))))))).....)))......... (-22.84 = -23.48 + 0.64)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:16:01 2006