| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,626,979 – 18,627,146 |
| Length | 167 |
| Max. P | 0.625935 |

| Location | 18,626,979 – 18,627,071 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.64 |
| Mean single sequence MFE | -22.47 |
| Consensus MFE | -13.75 |
| Energy contribution | -14.03 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.625935 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
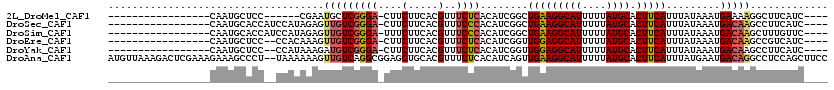
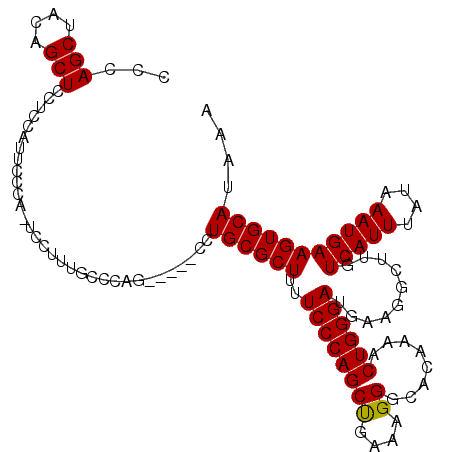
>2L_DroMel_CAF1 18626979 92 + 22407834 -----------------CAAUGCUCC------CGAAUGCUCGGGA-CUUCUUCACGUUUCUCACAUCGGCUGAAGGCAUUUUUAUGCACUUCAUUUAUAAAUGAAAAGGCUUCAUC---- -----------------.((((((((------(((....))))))-...(((((.(((.........))))))))))))).....((..((((((....))))))...))......---- ( -21.50) >DroSec_CAF1 104474 98 + 1 -----------------CAAUGCACCAUCCAUAGAGUUGUCGGGA-CUUCUUCACGUUUCCCACAUCGGCUGAAGGCAUUUUUAUGCACUUCAUUUAUAAAUGACAAGCCUUCAUC---- -----------------...............((..(((((((((-............))))........(((((((((....)))).))))).........)))))..)).....---- ( -19.00) >DroSim_CAF1 106082 98 + 1 -----------------CAAUGCACCAUCCAUAGAGUUGUCGGGA-UUUCUUCACGUUUCCCACAUCGGCUGAAGGCAUUUUUAUGCACUUCAUUUAUAAAUGACAAGCUUUGUUC---- -----------------.............(((((((((((((((-............))))........(((((((((....)))).))))).........))).))))))))..---- ( -23.80) >DroEre_CAF1 103469 96 + 1 -----------------CAAUGCUCC--CCACAAAGUUGUCGGGA-CUUCUUCACGUUUCUCACAUCGGUUGGAGGCAUUUUUAUGCACUUCAUUUAUAAAUGACAAGCCGUCAUC---- -----------------...((((((--(.(((....))).))))-.........((((((.((....)).))))))........)))............(((((.....))))).---- ( -19.40) >DroYak_CAF1 104885 96 + 1 -----------------CAAUGCUCC--CCAUAAAGAUGUCGGGA-CUUCUUCACGUUUCUCACAUCGGUUGGAGGCAUUUUUAUGCACUUCAUUUAUAAAUGACAAGCCUUCAUC---- -----------------.(((((..(--(.((...(((((.(..(-(........))..)..))))).)).))..))))).....((...(((((....)))))...)).......---- ( -20.70) >DroAna_CAF1 94535 118 + 1 AUGUUAAAGACUCGAAAGAAAGCCCU--UAAAAAAGUUGUCAGGCGGAGCUGCACGUUUCUCACAUCAGUUGAAGGCAUUUUUAUGCACUUCAUUUAUGAAUGACAGGCCUCCAGCUUCC ...........((....))..(((..--(((.....)))...)))(((((((...((((.(((..(((..(((((((((....)))).)))))....))).))).))))...))))))). ( -30.40) >consensus _________________CAAUGCUCC__CCAUAAAGUUGUCGGGA_CUUCUUCACGUUUCUCACAUCGGCUGAAGGCAUUUUUAUGCACUUCAUUUAUAAAUGACAAGCCUUCAUC____ ....................................(((((((((....(.....)..))))........(((((((((....)))).))))).........)))))............. (-13.75 = -14.03 + 0.28)
| Location | 18,627,035 – 18,627,146 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 85.69 |
| Mean single sequence MFE | -32.15 |
| Consensus MFE | -19.12 |
| Energy contribution | -18.88 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.59 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.533330 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
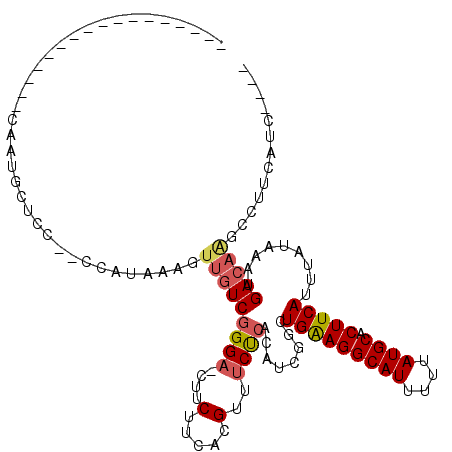
>2L_DroMel_CAF1 18627035 111 - 22407834 CCCAGCUACAGCUCCUCCAUUCCCA-UCCGCAUCCCAGCU---CCUGCGCUUUUCCCAGCUGAAGGGGCACAUAACUGGGAUGAAGCCUUUUCAUUUAUAAAUGAAGUGCAUAAA ...(((....)))............-....((((((((((---(((..(((......)))...)))))).......)))))))..((..(((((((....))))))).))..... ( -28.61) >DroSec_CAF1 104536 105 - 1 CCCAGCUACAGCUCCUCCAUUCCCA-UCCUGUGC---------CCUGCGCUUUUCCCAGCCGAAAGGGCACAAAACUGGGAUGAAGGCUUGUCAUUUAUAAAUGAAGUGCAUAAA ....((.((....(((.(((.((((-...(((((---------(((..(((......)))....))))))))....))))))).)))....(((((....))))).))))..... ( -32.10) >DroSim_CAF1 106144 105 - 1 CCCAGCUACAGCUCCUCCAUUCCCA-UCCUUUGC---------CCUGCGCUUUUCCCAGCCGAAAGGGCACAAAACUGGGAACAAAGCUUGUCAUUUAUAAAUGAAGUGCAUAAA ...(((....)))......((((((-.....(((---------(((..(((......)))....))))))......))))))....((...(((((....)))))...))..... ( -26.70) >DroEre_CAF1 103529 115 - 1 CCCAGCUACGGCUCCUCCAUGCCAGCUCCUUUGCCCAGCUUGCCCUGCGCUUUUCCCAGCUGGAGGGGCACAAAACUGGGAUGACGGCUUGUCAUUUAUAAAUGAAGUGCAUAAA (((((.....(((((((((.((.((((.........)))).)).....(((......))))))))))))......)))))......((...(((((....)))))...))..... ( -37.20) >DroYak_CAF1 104945 115 - 1 CCCAGCUACAGCUCCUCCGUCCCAGCUCCUUUGCCCAGGGUGCCCUGCGCUUUUCCCAGCUGGAUGGACACAAAACUGGGAUGAAGGCUUGUCAUUUAUAAAUGAAGUGCAUAAA (((((..........(((((((.((((.....((.((((....)))).)).......))))))))))).......)))))......((...(((((....)))))...))..... ( -36.13) >consensus CCCAGCUACAGCUCCUCCAUUCCCA_UCCUUUGCCCAG_____CCUGCGCUUUUCCCAGCUGAAAGGGCACAAAACUGGGAUGAAGGCUUGUCAUUUAUAAAUGAAGUGCAUAAA ...(((....)))................................((((((..((((((((....))........))))))..........(((((....))))))))))).... (-19.12 = -18.88 + -0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:15:58 2006