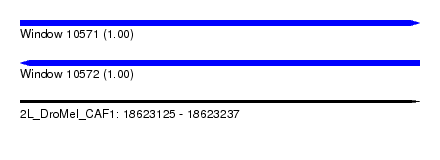
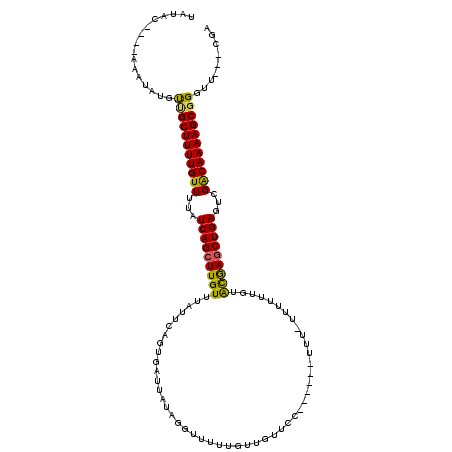
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,623,125 – 18,623,237 |
| Length | 112 |
| Max. P | 0.997050 |

| Location | 18,623,125 – 18,623,237 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.48 |
| Mean single sequence MFE | -19.70 |
| Consensus MFE | -13.07 |
| Energy contribution | -13.87 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.66 |
| SVM decision value | 2.57 |
| SVM RNA-class probability | 0.995421 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18623125 112 + 22407834 UCG---AACCCGCUUUUGUCGACUCAGCUCGUACAAAAAA-AUAUAUAUGGGAAAAACAAAAACCUAUAAUCACUGAAUAAACAAGCUGAUAAAACAAAAGCAACAUAUUU----GUAUA ...---.....((((((((....((((((.((........-.....((((((...........))))))............)).))))))....)))))))).(((....)----))... ( -22.21) >DroSec_CAF1 100705 106 + 1 UCG---AACCCGCUUUUGUCGACUCAGGUCGUACAAAAAA-AAA------GGAACAACAAAAACCUAUAAUCACUGAAUAAACAAGCUGAUAAAACAAAAGCAACGUAUUU----GUAUA ...---.....((((((((((((....)))).........-...------...................((((((.........)).))))...)))))))).........----..... ( -15.30) >DroSim_CAF1 102163 106 + 1 UCG---AACCCGCUUUUGUCGACUCAGCUCGUACAAAAAA-AAA------GGAACAACAAAAACCUAUAAUCACUGAAUAAACAAGCUGAUAAAACAAAAGCAGCAUAUUU----GUAUA ...---.....((((((((....((((((.((........-..(------((...........)))....((...))....)).))))))....)))))))).(((....)----))... ( -18.20) >DroEre_CAF1 99702 114 + 1 UCG---AACCCGCUUUUGUCGACUCAGCUUGUACACAGACUAGAAAAA-AGAAAAAA--CAAAUCUAUAAUCACUGAAUAAACAAGCUGAUAAAACAAAAGCGACUUACAUGUAUGUAUA ...---....(((((((((....(((((((((...(((..((((....-........--....))))......))).....)))))))))....)))))))))...((((....)))).. ( -24.79) >DroAna_CAF1 91515 97 + 1 UCGCGAGAGCCGCUUUUGCCGACUCAGCUCACGGAAUCA-----------G--GCAG--CAAACCCAUAAUCAUUGAAUAAACAAGCUGAUAAAACAAAAGCCACAUA--------AAUA ..((....)).(((((((.....((((((.......(((-----------(--....--..............)))).......)))))).....)))))))......--------.... ( -18.01) >consensus UCG___AACCCGCUUUUGUCGACUCAGCUCGUACAAAAAA_AAA______GGAACAACAAAAACCUAUAAUCACUGAAUAAACAAGCUGAUAAAACAAAAGCAACAUAUUU____GUAUA ...........((((((((....((((((.((.................................................)).))))))....)))))))).................. (-13.07 = -13.87 + 0.80)

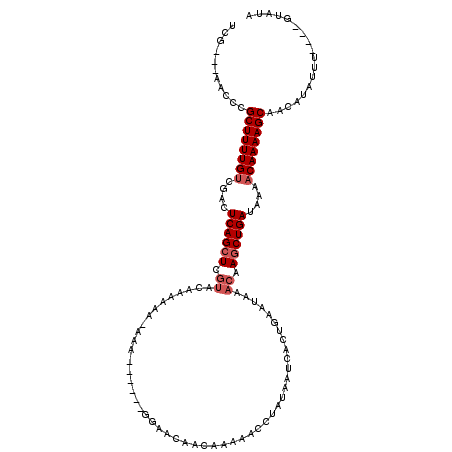
| Location | 18,623,125 – 18,623,237 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.48 |
| Mean single sequence MFE | -27.88 |
| Consensus MFE | -22.17 |
| Energy contribution | -21.65 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.80 |
| SVM decision value | 2.79 |
| SVM RNA-class probability | 0.997050 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18623125 112 - 22407834 UAUAC----AAAUAUGUUGCUUUUGUUUUAUCAGCUUGUUUAUUCAGUGAUUAUAGGUUUUUGUUUUUCCCAUAUAUAU-UUUUUUGUACGAGCUGAGUCGACAAAAGCGGGUU---CGA .....----.......(((((((((((...(((((((((......((((..(((.((...........)).)))..)))-).......)))))))))...)))))))))))...---... ( -26.72) >DroSec_CAF1 100705 106 - 1 UAUAC----AAAUACGUUGCUUUUGUUUUAUCAGCUUGUUUAUUCAGUGAUUAUAGGUUUUUGUUGUUCC------UUU-UUUUUUGUACGACCUGAGUCGACAAAAGCGGGUU---CGA .....----.......(((((((((((...((((.((((.....(((.((....(((..(.....)..))------)..-.)).))).)))).))))...)))))))))))...---... ( -21.20) >DroSim_CAF1 102163 106 - 1 UAUAC----AAAUAUGCUGCUUUUGUUUUAUCAGCUUGUUUAUUCAGUGAUUAUAGGUUUUUGUUGUUCC------UUU-UUUUUUGUACGAGCUGAGUCGACAAAAGCGGGUU---CGA .....----.......(((((((((((...(((((((((.....(((.((....(((..(.....)..))------)..-.)).))).)))))))))...)))))))))))...---... ( -30.40) >DroEre_CAF1 99702 114 - 1 UAUACAUACAUGUAAGUCGCUUUUGUUUUAUCAGCUUGUUUAUUCAGUGAUUAUAGAUUUG--UUUUUUCU-UUUUUCUAGUCUGUGUACAAGCUGAGUCGACAAAAGCGGGUU---CGA ..........((.((.(((((((((((...(((((((((.....(((.((..(.(((....--.....)))-.)..))....)))...)))))))))...))))))))))).))---)). ( -33.60) >DroAna_CAF1 91515 97 - 1 UAUU--------UAUGUGGCUUUUGUUUUAUCAGCUUGUUUAUUCAAUGAUUAUGGGUUUG--CUGC--C-----------UGAUUCCGUGAGCUGAGUCGGCAAAAGCGGCUCUCGCGA ....--------...((.(((((((((...((((((..(....(((.........(((...--..))--)-----------)))....)..))))))...))))))))).))........ ( -27.50) >consensus UAUAC____AAAUAUGUUGCUUUUGUUUUAUCAGCUUGUUUAUUCAGUGAUUAUAGGUUUUUGUUGUUCC______UUU_UUUUUUGUACGAGCUGAGUCGACAAAAGCGGGUU___CGA ................(((((((((((...(((((((((.................................................)))))))))...)))))))))))......... (-22.17 = -21.65 + -0.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:15:53 2006