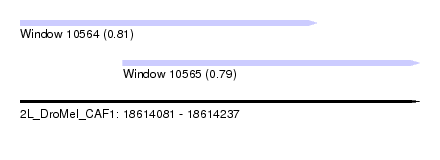
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,614,081 – 18,614,237 |
| Length | 156 |
| Max. P | 0.806784 |

| Location | 18,614,081 – 18,614,197 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 97.70 |
| Mean single sequence MFE | -31.85 |
| Consensus MFE | -28.67 |
| Energy contribution | -29.68 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.806784 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18614081 116 + 22407834 GUGCCGGCAAUAAAACGAAAUUGUUUUGAAUUAUUGAGAUCUGGCAGAAAGAGCGGCCUGUAACCGGAUGGUAACUUUCUGGUUUGGGCUUUUUUUCAUUUUGUUUAAAUAUAAUU .((((((((((((..((((.....))))..))))))....))))))(((((((.((((((.(((((((.(....)..))))))))))))))))))))................... ( -32.20) >DroSec_CAF1 91463 115 + 1 GUGCCGGCAAUAAAACGAAAUUGUUUUGAAUUAUUGAGAUCUGGCAGAAAGAGCGGCCUGUAACCGGAUGGUACCUUUCUGGUUUGGGCUUU-UUUCAUUUUGUUUAAAUAUAAUU .((((((((((((..((((.....))))..))))))....))))))((((((..((((((.(((((((.(....)..)))))))))))))))-))))................... ( -31.60) >DroSim_CAF1 92371 115 + 1 GUGCCGGCAAUAAAACGAAAUUGUUUUGAAUUAUUGAGAUCUGGCAGAAAGAGCGGCCUGUAACCGGAUGGUACCUUUCUGGUUUGGGCUUU-UUUCAUUUUGUUUAAAUAUAAUU .((((((((((((..((((.....))))..))))))....))))))((((((..((((((.(((((((.(....)..)))))))))))))))-))))................... ( -31.60) >DroEre_CAF1 90635 116 + 1 GUGCCGGCAAUAAAACGAAAUUGUUUUGAAUUAUUGAGAUCUGGCAGAAAGAGCGGCCUGUAACCGGAUGGUACCUUCCAGGUUUGGGCUUUUUUUCAUUUUGUUUGAAUAUAAUU .((((((((((((..((((.....))))..))))))....))))))(((((((.((((((.(((((((.(....).))).)))))))))))))))))................... ( -32.00) >consensus GUGCCGGCAAUAAAACGAAAUUGUUUUGAAUUAUUGAGAUCUGGCAGAAAGAGCGGCCUGUAACCGGAUGGUACCUUUCUGGUUUGGGCUUU_UUUCAUUUUGUUUAAAUAUAAUU .((((((((((((..((((.....))))..))))))....))))))(((((((.((((((.(((((((.((...)).))))))))))))))))))))................... (-28.67 = -29.68 + 1.00)


| Location | 18,614,121 – 18,614,237 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 95.54 |
| Mean single sequence MFE | -35.27 |
| Consensus MFE | -31.44 |
| Energy contribution | -32.25 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.02 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.792847 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18614121 116 + 22407834 CUGGCAGAAAGAGCGGCCUGUAACCGGAUGGUAACUUUCUGGUUUGGGCUUUUUUUCAUUUUGUUUAAAUAUAAUUUAUAUUUGGCGCAUAUAUAGCCAGGAUCACGGGACCCGGA (((((.(((((((.((((((.(((((((.(....)..))))))))))))))))))))....((((((((((((...))))))))).)))......))))).....(((...))).. ( -35.30) >DroSec_CAF1 91503 115 + 1 CUGGCAGAAAGAGCGGCCUGUAACCGGAUGGUACCUUUCUGGUUUGGGCUUU-UUUCAUUUUGUUUAAAUAUAAUUUAUAUUUGGCGCAUUUAUAGCCAGGACCUCGGGACCCGGA (((((.((((((..((((((.(((((((.(....)..)))))))))))))))-))))....((((((((((((...))))))))).)))......)))))....((((...)))). ( -34.80) >DroSim_CAF1 92411 115 + 1 CUGGCAGAAAGAGCGGCCUGUAACCGGAUGGUACCUUUCUGGUUUGGGCUUU-UUUCAUUUUGUUUAAAUAUAAUUUAUAUUUGGCGCAUUUAUAGCCAGGACUUCGGGACCCGGA (((((.((((((..((((((.(((((((.(....)..)))))))))))))))-))))....((((((((((((...))))))))).)))......)))))...(((((...))))) ( -35.30) >DroEre_CAF1 90675 116 + 1 CUGGCAGAAAGAGCGGCCUGUAACCGGAUGGUACCUUCCAGGUUUGGGCUUUUUUUCAUUUUGUUUGAAUAUAAUUUAUAUUUGGUGCAUUUAUAGCCAGGACCUCGGGACCCGGA (((((.(((((((.((((((.(((((((.(....).))).)))))))))))))))))....((((..((((((...))))))..).)))......)))))....((((...)))). ( -35.70) >consensus CUGGCAGAAAGAGCGGCCUGUAACCGGAUGGUACCUUUCUGGUUUGGGCUUU_UUUCAUUUUGUUUAAAUAUAAUUUAUAUUUGGCGCAUUUAUAGCCAGGACCUCGGGACCCGGA (((((.(((((((.((((((.(((((((.((...)).))))))))))))))))))))....((((((((((((...))))))))).)))......))))).....(((...))).. (-31.44 = -32.25 + 0.81)

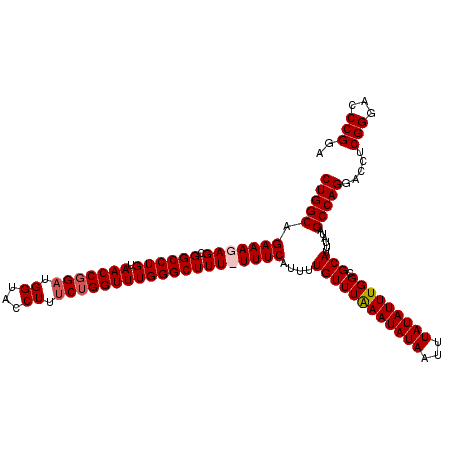
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:15:46 2006