| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,613,476 – 18,613,570 |
| Length | 94 |
| Max. P | 0.715893 |

| Location | 18,613,476 – 18,613,570 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.29 |
| Mean single sequence MFE | -28.02 |
| Consensus MFE | -9.27 |
| Energy contribution | -10.27 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.73 |
| Structure conservation index | 0.33 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.715893 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
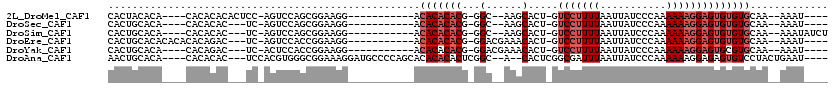
>2L_DroMel_CAF1 18613476 94 - 22407834 CACUACACA----CACACACACUCC-AGUCCAGCGGAAGG-----------ACACACACG-GGC--AAGCACU-GUCCUUUUAAUUAUCCCAAAAAAGGAGUGUGUGCAA--AAAU---- .........----..((((((((((-.((((.......))-----------))......(-(((--(.....)-))))...................))))))))))...--....---- ( -30.10) >DroSec_CAF1 90905 91 - 1 CACUGCACA----CACACAC---UC-AGUCCAGCGGAAGG-----------ACACACACG-GGC--AAGCACU-GUCCUUUUAAUUAUCCCAAAAAAGGAGUGUGUGCAA--AAAU---- ...((((((----(((....---..-.((((.......))-----------))......(-(((--(.....)-)))).........(((.......)))))))))))).--....---- ( -28.30) >DroSim_CAF1 91796 95 - 1 CACUGCACA----CACACAC---UC-AGUCCAGCGGAAGG-----------ACACACACG-GGC--AAGCACU-GUCCUUUUAAUUAUCCCAAAAAAGGAGUGUGUGCAA--AAAUAUCU ...((((((----(((....---..-.((((.......))-----------))......(-(((--(.....)-)))).........(((.......)))))))))))).--........ ( -28.30) >DroEre_CAF1 90020 97 - 1 CACUGCACACACACACAGAC---UC-AGUCCACCGGAAGG-----------ACACACACG-GGACGAAACACU-GUCCUUUUAAUUAUCCCAAAAAAGGAGUGUGUGCAA--AAAU---- ...(((((((((...(((..---..-.((((.((....))-----------.(......)-))))......))-)(((((((...........)))))))))))))))).--....---- ( -31.70) >DroYak_CAF1 90961 93 - 1 CACUGCACA----CACAGAC---UC-ACUCCACCGGAAGG-----------ACACACACG-GGACGAAACACU-GUCCUUUUAAUUAUCCCAAAAAAGGAGUGCGUGCAA--AAAU---- ...(((((.----.......---.(-(((((.((....))-----------........(-(((((......)-)))))..................)))))).))))).--....---- ( -25.00) >DroAna_CAF1 83503 105 - 1 AACUGCACA----CACACAC---UCCACGUGGGCGGAAAGGAUGCCCCAGCACACACACUCGGC--A--CACUCGGCGAUUUAAUUAUCCCAAAAAAGGAGAGUGUCCUACUGAAU---- .........----.((((.(---(((..(((.((.((..(..(((....)))..)....)).))--.--)))..((.(((......)))))......)))).))))..........---- ( -24.70) >consensus CACUGCACA____CACACAC___UC_AGUCCAGCGGAAGG___________ACACACACG_GGC__AAGCACU_GUCCUUUUAAUUAUCCCAAAAAAGGAGUGUGUGCAA__AAAU____ ....................................................(((((((...(......).....(((((((...........))))))))))))))............. ( -9.27 = -10.27 + 1.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:15:44 2006