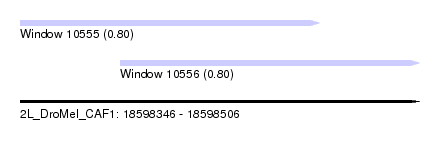
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,598,346 – 18,598,506 |
| Length | 160 |
| Max. P | 0.803655 |

| Location | 18,598,346 – 18,598,466 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.17 |
| Mean single sequence MFE | -41.95 |
| Consensus MFE | -26.29 |
| Energy contribution | -27.10 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.803655 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
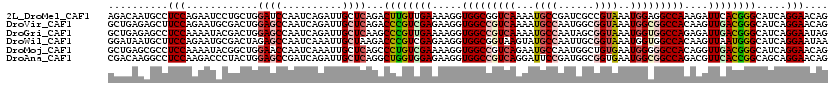
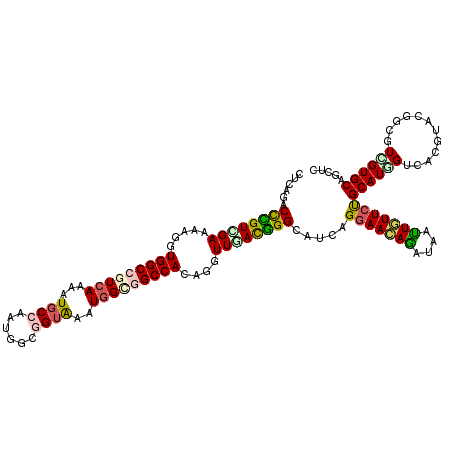
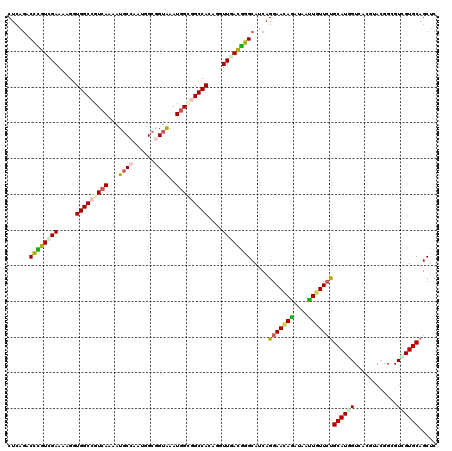
>2L_DroMel_CAF1 18598346 120 + 22407834 AGACAAUGCCUCCAGAAUCCUGCUGGAUCCAAUCAGAUUGCUCAGACUUGUUGAAAAGGUGGCGGUCAAAAUGCCGAUCGCCGUAAAUGGAGGCCAAAGAUUCACGGGCAUCAGGAACAG .....((((((...(((((....(((.((((..............((((.......))))(((((((........))))))).....))))..)))..)))))..))))))......... ( -33.00) >DroVir_CAF1 116721 120 + 1 GCUGAGAGCUUCCAGAAUGCGACUGGAGCCAAUCAGAUUGCUCAGACCCGUCGAGAAGGUGGCCGUCAAAAUGCCAAUGGCGGUAAAUGGCGGCCACAAGUUGACGGGCAUCAGGAACAG .((((((((((((((.......))))))...........)))).(.((((((((....((((((((((...((((......))))..))))))))))...))))))))).))))...... ( -51.60) >DroGri_CAF1 90612 120 + 1 GCUGAGAGCCUCCAAAAUACGACUGGAGCCAAUCAGAUUGCUCAAGCCCGUUGAAAAGGUGGCCGUCAAAAUGCCAAUAGCGGUAAAUGGUGGCCAGAGAUUGACGGGCAUCAGGAAUAG .(((((((((((((.........)))))...........))))..((((((..(.....(((((..((...((((......))))..))..)))))....)..)))))).))))...... ( -43.80) >DroWil_CAF1 155843 120 + 1 GGAUAAUGCUUCCAGAAUGCGACUAGAGCCAAUCAAAUUGCUAAGACCCGUCGAGAAGGUGGCGGUAAGUAUGCCAAUUGCGGUAAAUGGUGGCCACAAGUUAAUGGGCAUCAGGAAUAA .........((((.((..((((...((.....))...))))...(.(((((..(....(((((.(...((.((((......)))).))..).)))))...)..)))))).)).))))... ( -27.70) >DroMoj_CAF1 110480 120 + 1 GCUGAGCGCCUCCAAAAUACGGCUGGAACCAAUCAAAUUGCUCAGCCCUGUCGAAAAGGUGGCCGUCAGAAUGCCAAUGGCUGUGAAUGGGGGCCACAGGUUGACGGGCAUCAGGAACAG ((((((((((..........)))((....))........)))))))((((((((....((((((.(((....(((...)))......))).))))))...))))))))............ ( -47.40) >DroAna_CAF1 69484 120 + 1 CGACAAGGCCUCCAAGACCCUACUGGAGCCGAUCAGAUUGCUCAGGCUGGUGGAGAAGGUGGCCGUCAGGAUUCCGAUGGCGGUGAAUGGCGGCCAGACGUUCACCGGCAGCAGGAACAG .((...(((.((((.........)))))))..))....((.((..((((.(((.(((.((((((((((.....(((....)))....))))))))..)).))).))).))))..)).)). ( -48.20) >consensus GCAGAAAGCCUCCAAAAUACGACUGGAGCCAAUCAGAUUGCUCAGACCCGUCGAAAAGGUGGCCGUCAAAAUGCCAAUGGCGGUAAAUGGCGGCCACAGGUUGACGGGCAUCAGGAACAG ..........(((............((((..........))))...((((((((.....(((((((((...((((......))))..)))))))))....)))))))).....))).... (-26.29 = -27.10 + 0.81)
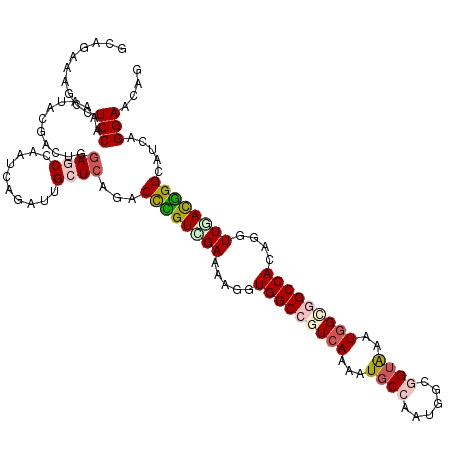
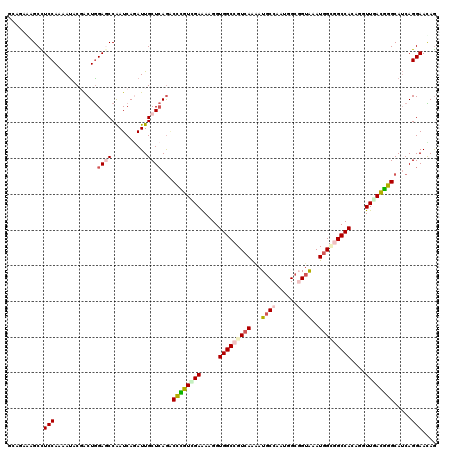
| Location | 18,598,386 – 18,598,506 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.67 |
| Mean single sequence MFE | -43.70 |
| Consensus MFE | -30.64 |
| Energy contribution | -30.98 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.800921 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18598386 120 + 22407834 CUCAGACUUGUUGAAAAGGUGGCGGUCAAAAUGCCGAUCGCCGUAAAUGGAGGCCAAAGAUUCACGGGCAUCAGGAACAGGUAAUUGUUCUGCAUGGUAAAGUACGGAGUCGUGCAGCUC (((..((((.......))))(((((((........))))))).......))).............(((((((((((((((....)))))))...))))...(((((....))))).)))) ( -33.00) >DroVir_CAF1 116761 120 + 1 CUCAGACCCGUCGAGAAGGUGGCCGUCAAAAUGCCAAUGGCGGUAAAUGGCGGCCACAAGUUGACGGGCAUCAGGAACAGAUAAUUGUUCUGCAUUGUCUGGUACGGUGUGGUGCAGCUC .......(((((((....((((((((((...((((......))))..))))))))))...)))))))(((((.(((((((....)))))))(((((((.....))))))))))))..... ( -51.40) >DroGri_CAF1 90652 120 + 1 CUCAAGCCCGUUGAAAAGGUGGCCGUCAAAAUGCCAAUAGCGGUAAAUGGUGGCCAGAGAUUGACGGGCAUCAGGAAUAGAUAACUAUUGCGCAUCGUCACCGACGGCGUCGUGCAGCUC .....((((((..(.....(((((..((...((((......))))..))..)))))....)..))))))......(((((....)))))((((..((((......))))..))))..... ( -43.90) >DroWil_CAF1 155883 120 + 1 CUAAGACCCGUCGAGAAGGUGGCGGUAAGUAUGCCAAUUGCGGUAAAUGGUGGCCACAAGUUAAUGGGCAUCAGGAAUAAGUAAUUCUUCUGCAUGGUGGCAAACGGAGUCGUGCAGCUC ....(((((((((......))))))...((.(((((..(((((....(((((.(((........))).)))))(((((.....))))).)))))...))))).))...)))......... ( -37.30) >DroMoj_CAF1 110520 120 + 1 CUCAGCCCUGUCGAAAAGGUGGCCGUCAGAAUGCCAAUGGCUGUGAAUGGGGGCCACAGGUUGACGGGCAUCAGGAACAGAUAGUUGUUCUGCAUGGUCACAUACGGCGUGGUGCAGCUC ...(((.(((((((....((((((.(((....(((...)))......))).))))))...)))))))(((((((((((((....))))))).....(((......))).)))))).))). ( -45.90) >DroAna_CAF1 69524 120 + 1 CUCAGGCUGGUGGAGAAGGUGGCCGUCAGGAUUCCGAUGGCGGUGAAUGGCGGCCAGACGUUCACCGGCAGCAGGAACAGGUAGUUGUUCUGCAUCGUGAAGUAGGGCGUGGUGCAGCAC .....(((((((((...(.(((((((((.....(((....)))....)))))))))..).))))))))).((.(((((((....)))))))((((((((........)))))))).)).. ( -50.70) >consensus CUCAGACCCGUCGAAAAGGUGGCCGUCAAAAUGCCAAUGGCGGUAAAUGGCGGCCACAGGUUGACGGGCAUCAGGAACAGAUAAUUGUUCUGCAUGGUCACGUACGGCGUCGUGCAGCUC ......((((((((.....(((((((((...((((......))))..)))))))))....)))))))).....(((((((....)))))))((((((............))))))..... (-30.64 = -30.98 + 0.34)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:15:37 2006