
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,592,658 – 18,592,754 |
| Length | 96 |
| Max. P | 0.801152 |

| Location | 18,592,658 – 18,592,754 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 72.81 |
| Mean single sequence MFE | -37.55 |
| Consensus MFE | -14.15 |
| Energy contribution | -13.96 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.47 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.38 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.801152 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18592658 96 - 22407834 ACCUUC---CAGCAGUUGGAGCAGCUCAUGUACCAGCAG---------GAGUUCA------GCACCAGCGACAGCCAGUCGGAUGGCGCCAACAGUUGCUCCUUGGAG---AUGUAU ...(((---(((.....(((((((((......((....)---------).(((..------((.(((.((((.....))))..))).)).))))))))))))))))))---...... ( -32.00) >DroVir_CAF1 105744 111 - 1 AAUUUGCAGGAUCAGCUGGAGCAGCUGAUGAACCAGCAGCAGGAUAGGGAUUUUAGUG---GCAGCGCGGACAGUCUGUCCGAUGGCACCAAUAGCUGCUCCCUGGAG---CUGUAC ....(((((..((((..((((((((((.((..((.......))............(((---.((...(((((.....))))).)).))))).))))))))))))))..---))))). ( -43.40) >DroPse_CAF1 109207 102 - 1 GCCUUU---CAGCAGUUGGAGCAGUUCAUGUACCAACAG---------GAGCUCUGUG---GCAGCGGCGACAGUCUCUCGGAUGAGACCAACAGUUGUUCCAUGGAGCAGCUAUAC ......---.(((.(((((.(((.....))).)))))..---------..((((((((---(.((((((....(((((......))))).....))))))))))))))).))).... ( -41.10) >DroWil_CAF1 148523 102 - 1 GCGUUC---CAACAGCUGGAGCAGUUCAUGUACCAGCAG---------GAUUUCAAUGGCUGCAGCGGAGAUAGUCUCUCGGAUGGCACCAACAGUAGUUCCCUCGAA---AUGUAC (((((.---(....(((((.(((.....))).))))).(---------((((....(((.(((..(.((((.....)))).)...)))))).....)))))....).)---)))).. ( -28.90) >DroAna_CAF1 64134 102 - 1 GCCUUC---CAGCAACUGGAGCAACUGAUAUACCAACAG---------GACUUUACCU---GCAACGCCGACAGCCUCUCCGAGGGCACCAGCAGCUGCUCCAUGGAGCAGUUGUAC ((((((---........((((...(((.........(((---------(......)))---).........)))...))))))))))....((((((((((....)))))))))).. ( -36.47) >DroPer_CAF1 107807 102 - 1 GCCUUU---CAGCAGUUGGAGCAGUUCAUGUACCAACAG---------GAGCUCUGUG---GCAGCGGCGACAGUCUCUCGGAUGAGACCAACAGUUGUUCCAUGGAGCAGCUGUAC ......---((((.(((((.(((.....))).)))))..---------..((((((((---(.((((((....(((((......))))).....))))))))))))))).))))... ( -43.40) >consensus GCCUUC___CAGCAGCUGGAGCAGCUCAUGUACCAACAG_________GAGUUCAGUG___GCAGCGGCGACAGUCUCUCGGAUGGCACCAACAGUUGCUCCAUGGAG___CUGUAC ..............(((((.(((.....))).)))))..............(((.......(((((.......(((........))).......)))))......)))......... (-14.15 = -13.96 + -0.19)
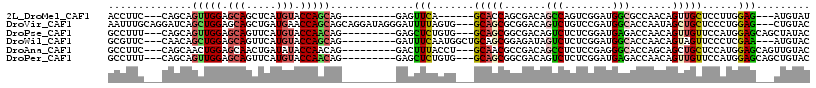

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:15:35 2006