| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,585,370 – 18,585,467 |
| Length | 97 |
| Max. P | 0.882505 |

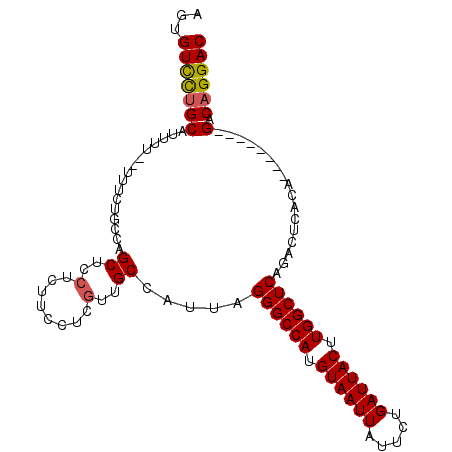
| Location | 18,585,370 – 18,585,467 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 88.65 |
| Mean single sequence MFE | -29.55 |
| Consensus MFE | -24.09 |
| Energy contribution | -24.07 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.862622 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18585370 97 + 22407834 GUCCUGUC--------UGUAAGUCUGAGCCAAGUAAUCAGAAUAAUUACAUGGCCCUAAUGGCAACGAGGAAGAGGAGCUGGCAGAAAAAAAAAUGCAGGACACU ((((((((--------(((.(((((..((((.(((((.......))))).))))(((...(....).)))....))).)).))))..........)))))))... ( -29.10) >DroSec_CAF1 61782 95 + 1 GUCCUGUC--------UGUGAGUCUGAGCCAAGUAAUCAGAAUAAUUACAUGGCCCUAAUGGCAACGAGGAAGAGGAGCUGGCAGAAA--AAAAUGCAGGACACU ((((((((--------(((.(((((..((((.(((((.......))))).))))(((...(....).)))....))).)).))))...--.....)))))))... ( -31.40) >DroSim_CAF1 62680 95 + 1 GUCCUGUC--------UGUGAGUCUGAGCCAAGUAAUCAGAAUAAUUACAUGGCCCUAAUGGCAACGAGGAAGAGGAGCUGGCAGAAA--AAAAUGCAGGACACU ((((((((--------(((.(((((..((((.(((((.......))))).))))(((...(....).)))....))).)).))))...--.....)))))))... ( -31.40) >DroEre_CAF1 61410 91 + 1 GUCCUGUC--------UGUGAGUCUGAGCCAAGUAAUCAGAAUAAUUACAUGGCCCUAAUGGCCACGGGGAAGAGGAGCUGGCA---A---AAAUGCAGGACACU (((((((.--------(((.(((((...((..(((((.......))))).(((((.....)))))...))....))).)).)))---.---....)))))))... ( -29.40) >DroYak_CAF1 62267 100 + 1 GUCCUGUCUGCGAGUCUGUGAGUCUGAGCCAAGUAAUCAGAAUAAUUACAUGGCCCUAAUGGCCACGAGGAAGAGGAGCUGGUA---A--AAAAUGCAGGACUCU (((((((.(((.(((((((((.(((((.........)))))....)))).(((((.....))))).........))).)).)))---.--.....)))))))... ( -30.10) >DroAna_CAF1 56749 97 + 1 GUCCUGUC--------UGUGAGCCUGAGCCAAGUAAUCAGAAUAAUUACAUGGCCCUAAUGGCAAUGAGGAAGAGGCGCCGGCGAAAAGGAAAACGCCAAACAGU .......(--------(((..((((..((((.(((((.......))))).))))(((.((....)).)))...))))...((((..........))))..)))). ( -25.90) >consensus GUCCUGUC________UGUGAGUCUGAGCCAAGUAAUCAGAAUAAUUACAUGGCCCUAAUGGCAACGAGGAAGAGGAGCUGGCAGAAA__AAAAUGCAGGACACU (((((((.........(((.(((((..((((.(((((.......))))).))))(((...(....).)))....)).))).)))...........)))))))... (-24.09 = -24.07 + -0.03)

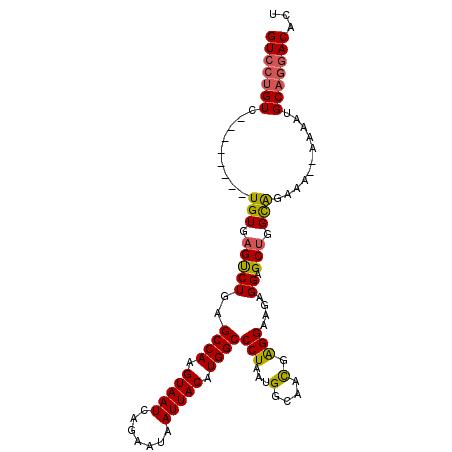

| Location | 18,585,370 – 18,585,467 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 88.65 |
| Mean single sequence MFE | -26.77 |
| Consensus MFE | -22.21 |
| Energy contribution | -22.27 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.882505 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18585370 97 - 22407834 AGUGUCCUGCAUUUUUUUUUCUGCCAGCUCCUCUUCCUCGUUGCCAUUAGGGCCAUGUAAUUAUUCUGAUUACUUGGCUCAGACUUACA--------GACAGGAC ...(((((((...........((.((((...........)))).))...((((((.((((((.....)))))).)))))).........--------).)))))) ( -25.80) >DroSec_CAF1 61782 95 - 1 AGUGUCCUGCAUUUU--UUUCUGCCAGCUCCUCUUCCUCGUUGCCAUUAGGGCCAUGUAAUUAUUCUGAUUACUUGGCUCAGACUCACA--------GACAGGAC ...(((((((.....--....((.((((...........)))).))...((((((.((((((.....)))))).)))))).........--------).)))))) ( -25.80) >DroSim_CAF1 62680 95 - 1 AGUGUCCUGCAUUUU--UUUCUGCCAGCUCCUCUUCCUCGUUGCCAUUAGGGCCAUGUAAUUAUUCUGAUUACUUGGCUCAGACUCACA--------GACAGGAC ...(((((((.....--....((.((((...........)))).))...((((((.((((((.....)))))).)))))).........--------).)))))) ( -25.80) >DroEre_CAF1 61410 91 - 1 AGUGUCCUGCAUUU---U---UGCCAGCUCCUCUUCCCCGUGGCCAUUAGGGCCAUGUAAUUAUUCUGAUUACUUGGCUCAGACUCACA--------GACAGGAC ...(((((((....---.---.((((..............)))).....((((((.((((((.....)))))).)))))).........--------).)))))) ( -26.84) >DroYak_CAF1 62267 100 - 1 AGAGUCCUGCAUUUU--U---UACCAGCUCCUCUUCCUCGUGGCCAUUAGGGCCAUGUAAUUAUUCUGAUUACUUGGCUCAGACUCACAGACUCGCAGACAGGAC ...(((((((.....--.---.....((..(........)..)).....((((((.((((((.....)))))).)))))).................).)))))) ( -25.90) >DroAna_CAF1 56749 97 - 1 ACUGUUUGGCGUUUUCCUUUUCGCCGGCGCCUCUUCCUCAUUGCCAUUAGGGCCAUGUAAUUAUUCUGAUUACUUGGCUCAGGCUCACA--------GACAGGAC .((((((((((((..(......)..))))))...........(((....((((((.((((((.....)))))).)))))).)))....)--------)))))... ( -30.50) >consensus AGUGUCCUGCAUUUU__UUUCUGCCAGCUCCUCUUCCUCGUUGCCAUUAGGGCCAUGUAAUUAUUCUGAUUACUUGGCUCAGACUCACA________GACAGGAC ...(((((((................((..(........)..)).....((((((.((((((.....)))))).)))))).................).)))))) (-22.21 = -22.27 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:15:32 2006