
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,581,855 – 18,581,995 |
| Length | 140 |
| Max. P | 0.939861 |

| Location | 18,581,855 – 18,581,960 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 76.95 |
| Mean single sequence MFE | -26.84 |
| Consensus MFE | -15.67 |
| Energy contribution | -16.12 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939861 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
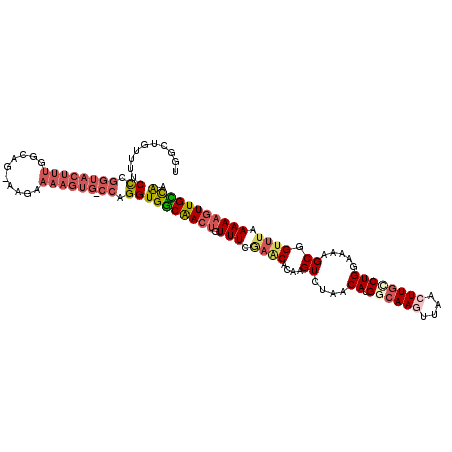
>2L_DroMel_CAF1 18581855 105 - 22407834 GGCCAGGUGGCAACUGUUUCGAACACAAGUCUAACAUGGCAAGUUAACUUGCCUGGAAAAGCGGUUUUAAAAGUUGCUAAAAAGUUGUAUUUACA---AUU-AUGUAUA .......((((((((.(((.((((.....((((....((((((....))))))))))......)))).)))))))))))...((((((....)))---)))-....... ( -26.40) >DroSec_CAF1 58284 105 - 1 G-CCAGGUGGCAACUGUUUCGAACGCAAGUCUAACAUGGCAAUUUAACUUGCCUGGAAAAGCGGUUUAAAAAGUUGCUAAAAAGUUGUAUUUACA---AGUGACAUAAA .-.....((((((((.(((..(((((...((((....(((((......)))))))))...)).)))..)))))))))))....((..(.......---.)..))..... ( -24.70) >DroSim_CAF1 59169 105 - 1 G-CCAGGUGGCAACUGUUUCGAACACAAGUCUAACAAGGCAAGUUAACUUGCCUGGAAAAGCGGUUUAAAAAGUUGCUAGCAAGUUGUAUUUACA---AGUGACAUACA .-...(.((((((((.(((..(((....((......(((((((....)))))))......)).)))..))))))))))).)....((((((((..---..))).))))) ( -29.50) >DroEre_CAF1 57891 98 - 1 G-CCAGGUGGCAACUGUUUUAAACAGAAGUCAAACAUGGCAAGUUAACUUGACUGGAAAAGCGGCUUAAAAUGUUGUCAAAGAGUUGUAUUUGCAGAAA---------- (-(((..((((..((((.....))))..))))....))))............(((.(((.(((((((..............))))))).))).)))...---------- ( -25.84) >DroYak_CAF1 58658 108 - 1 G-CCAGGUGGCAACUGUUUUGAACACAAGUCCAACAUGGCAAGUUAACUUGUCUGGAAAAGCGGUUUAAAAAGUUGUCAAAAAGUUGUAUUUACAGAAAGAGCCACACA .-...((((((((((.((((((((.....((((....((((((....))))))))))......)))))))))))))))......((((....)))).....)))..... ( -29.60) >DroAna_CAF1 51998 97 - 1 G-UCAGGUGACGAGUGUUUAGGGCACAUGUCGAACAGGGAAAGUUUGGUUAACUGGAAAAGCUGUG-GAAAAGUUGUCGGAAACUAGAACUGACUG----------CCA (-((((.(((((.((((.....)))).)))))...........(((((((..((((...((((...-....)))).)))).))))))).)))))..----------... ( -25.00) >consensus G_CCAGGUGGCAACUGUUUCGAACACAAGUCUAACAUGGCAAGUUAACUUGCCUGGAAAAGCGGUUUAAAAAGUUGCCAAAAAGUUGUAUUUACA___AGU_ACAUACA .......((((((((.(((.((((....((....((.((((((....)))))))).....)).)))).))))))))))).............................. (-15.67 = -16.12 + 0.45)



| Location | 18,581,880 – 18,581,995 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 81.51 |
| Mean single sequence MFE | -34.30 |
| Consensus MFE | -18.43 |
| Energy contribution | -19.97 |
| Covariance contribution | 1.53 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.588549 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18581880 115 - 22407834 UGGCUGUUUUCUCGGUACUUUGGCAG-AAGAAAAGUGGCCAGGUGGCAACUGUUUCGAACACAAGUCUAACAUGGCAAGUUAACUUGCCUGGAAAAGCGGUUUUAAAAGUUGCUAA (((((((((((((.((......)).)-.)))))).))))))..((((((((.(((.((((.....((((....((((((....))))))))))......)))).))))))))))). ( -35.40) >DroSec_CAF1 58310 114 - 1 UGGCUGUUUUCCCGGUACUUUGGCAG-AAGAAAAGUG-CCAGGUGGCAACUGUUUCGAACGCAAGUCUAACAUGGCAAUUUAACUUGCCUGGAAAAGCGGUUUAAAAAGUUGCUAA ..........((.((((((((.....-....))))))-)).))((((((((.(((..(((((...((((....(((((......)))))))))...)).)))..))))))))))). ( -34.70) >DroSim_CAF1 59195 114 - 1 UGGCUGUUUUCCCGGUACUUUGGCAG-AAGAAAAGUG-CCAGGUGGCAACUGUUUCGAACACAAGUCUAACAAGGCAAGUUAACUUGCCUGGAAAAGCGGUUUAAAAAGUUGCUAG ..........((.((((((((.....-....))))))-)).))((((((((.(((..(((....((......(((((((....)))))))......)).)))..))))))))))). ( -36.90) >DroEre_CAF1 57910 113 - 1 UGGCUGUUUUCUCGGUACUUUAGCCA--AGAAAAGUG-CCAGGUGGCAACUGUUUUAAACAGAAGUCAAACAUGGCAAGUUAACUUGACUGGAAAAGCGGCUUAAAAUGUUGUCAA .(((((((((..((((........((--((.((..((-(((..((((..((((.....))))..))))....)))))..))..))))))))..))))))))).............. ( -33.20) >DroYak_CAF1 58687 114 - 1 UGGCUGUUUUCUCGGUACUUUAGCAA-AAGAAAAGUG-CCAGGUGGCAACUGUUUUGAACACAAGUCCAACAUGGCAAGUUAACUUGUCUGGAAAAGCGGUUUAAAAAGUUGUCAA ((((..((((((..((......))..-.))))))..)-)))..((((((((.((((((((.....((((....((((((....))))))))))......)))))))))))))))). ( -37.10) >DroAna_CAF1 52017 104 - 1 UAGCUCCUUUCC----------GCCGAGGGAGGAGUG-UCAGGUGACGAGUGUUUAGGGCACAUGUCGAACAGGGAAAGUUUGGUUAACUGGAAAAGCUGUG-GAAAAGUUGUCGG ..((((((..(.----------.....)..))))))(-.(((.(((((.((((.....)))).))))...((.((....((..(....)..))....)).))-....).))).).. ( -28.50) >consensus UGGCUGUUUUCCCGGUACUUUGGCAG_AAGAAAAGUG_CCAGGUGGCAACUGUUUCGAACACAAGUCUAACAUGGCAAGUUAACUUGCCUGGAAAAGCGGUUUAAAAAGUUGCCAA ..........((.((((((((..........)))))).)).))((((((((.(((.((((....((....((.((((((....)))))))).....)).)))).))))))))))). (-18.43 = -19.97 + 1.53)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:15:29 2006