| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,535,118 – 18,535,216 |
| Length | 98 |
| Max. P | 0.996556 |

| Location | 18,535,118 – 18,535,216 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 89.30 |
| Mean single sequence MFE | -26.47 |
| Consensus MFE | -21.72 |
| Energy contribution | -21.88 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.60 |
| SVM RNA-class probability | 0.995640 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18535118 98 + 22407834 UUAAUUACGCUCCAAUUGGCAGACAGGCCGAAGAGAAACUGCCAAAAAAAAAAAAAAUGGGAAAAUGUGGCAGUAGUGGCCAGGGCGGAAGCUGGGAA ....((((.(((...(((((......))))).)))..(((((((.......................))))))).))))((..(((....))).)).. ( -25.80) >DroSec_CAF1 12353 91 + 1 UUAAUUACGCUCCAAUUGGCAGACAGGCCGAAGAAAAACUGCCGAA-------AAAAUGGGAAAAUGUGGCAGUGGCCACGGGGGCGGAAGCUGGGAA .......((((((..(((((((................))))))).-------............((((((....))))))))))))........... ( -28.69) >DroSim_CAF1 12346 91 + 1 UUAAUUACGCUCCAAUUGGCAGACAGGCCGAAGAAAAACUGCCGAA-------AAAAUGGGAAAAUGUGGCAGUGGCCACGGGGGCGGAAGCUGGGAA .......((((((..(((((((................))))))).-------............((((((....))))))))))))........... ( -28.69) >DroEre_CAF1 12515 88 + 1 UUAAUUACGCUCCAAUUGGCAGACAGGCCGAGGAAAAAGUGCCGAA-------AAAAUGGGAAAAUGUGGCAGCGGCCA---GAGCGGAAGCUGGGAA .......((((((..(((((......)))))))).....(((((..-------...((......)).)))))))).((.---.(((....))).)).. ( -25.30) >DroYak_CAF1 12377 88 + 1 UUAAUUACGCUCCAAUUGGCAGACAGGCCGAAGAAAAACUGCCGAA-------AAAAUAGGAAAAUGUGGCAGCGGCCA---UGGCGGAAGCUGGGAA ..........(((............(((((........((((((..-------..............))))))))))).---.(((....))).))). ( -23.89) >consensus UUAAUUACGCUCCAAUUGGCAGACAGGCCGAAGAAAAACUGCCGAA_______AAAAUGGGAAAAUGUGGCAGUGGCCAC__GGGCGGAAGCUGGGAA ..........(((..(((((((.....(....).....))))))).....................(((((....)))))...(((....))).))). (-21.72 = -21.88 + 0.16)

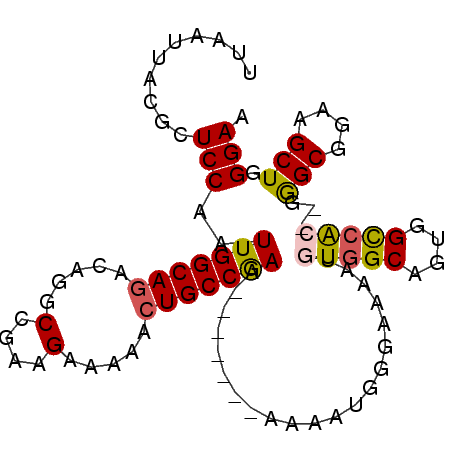

| Location | 18,535,118 – 18,535,216 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 89.30 |
| Mean single sequence MFE | -22.07 |
| Consensus MFE | -18.76 |
| Energy contribution | -19.96 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.71 |
| SVM RNA-class probability | 0.996556 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18535118 98 - 22407834 UUCCCAGCUUCCGCCCUGGCCACUACUGCCACAUUUUCCCAUUUUUUUUUUUUUUGGCAGUUUCUCUUCGGCCUGUCUGCCAAUUGGAGCGUAAUUAA ......(((((.....((((.......))))......................(((((((.....(....).....)))))))..)))))........ ( -21.80) >DroSec_CAF1 12353 91 - 1 UUCCCAGCUUCCGCCCCCGUGGCCACUGCCACAUUUUCCCAUUUU-------UUCGGCAGUUUUUCUUCGGCCUGUCUGCCAAUUGGAGCGUAAUUAA ......(((((.......(((((....))))).............-------...(((((.....(....).....)))))....)))))........ ( -24.90) >DroSim_CAF1 12346 91 - 1 UUCCCAGCUUCCGCCCCCGUGGCCACUGCCACAUUUUCCCAUUUU-------UUCGGCAGUUUUUCUUCGGCCUGUCUGCCAAUUGGAGCGUAAUUAA ......(((((.......(((((....))))).............-------...(((((.....(....).....)))))....)))))........ ( -24.90) >DroEre_CAF1 12515 88 - 1 UUCCCAGCUUCCGCUC---UGGCCGCUGCCACAUUUUCCCAUUUU-------UUCGGCACUUUUUCCUCGGCCUGUCUGCCAAUUGGAGCGUAAUUAA ...........(((((---..(....((((...............-------...))))..........(((......)))..)..)))))....... ( -17.77) >DroYak_CAF1 12377 88 - 1 UUCCCAGCUUCCGCCA---UGGCCGCUGCCACAUUUUCCUAUUUU-------UUCGGCAGUUUUUCUUCGGCCUGUCUGCCAAUUGGAGCGUAAUUAA ......(((((.....---((((....))))..............-------...(((((.....(....).....)))))....)))))........ ( -21.00) >consensus UUCCCAGCUUCCGCCC__GUGGCCACUGCCACAUUUUCCCAUUUU_______UUCGGCAGUUUUUCUUCGGCCUGUCUGCCAAUUGGAGCGUAAUUAA ......(((((.......(((((....))))).......................(((((.....(....).....)))))....)))))........ (-18.76 = -19.96 + 1.20)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:15:19 2006