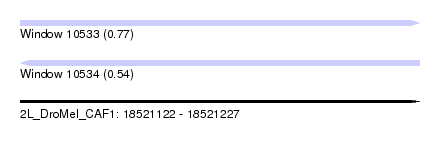
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,521,122 – 18,521,227 |
| Length | 105 |
| Max. P | 0.767694 |

| Location | 18,521,122 – 18,521,227 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.31 |
| Mean single sequence MFE | -37.88 |
| Consensus MFE | -28.35 |
| Energy contribution | -29.43 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.767694 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18521122 105 + 22407834 AACUUAUGUGUGCCAAUUGCC---UGCCGUCCGUUGCCUGCAUAUUGGCCAAUCGUUAUGGAUUUGACUGGCCUCAAUGGGCAGC---CACGCACCUGGCAA---------AUGGGUAAU .((((((...(((((..(((.---.((.((((((((..........(((((.(((.........))).))))).)))))))).))---...)))..))))).---------))))))... ( -37.40) >DroPse_CAF1 50225 111 + 1 A-CUUCAGUGGGCCAAUUGGCAAUUGCCGCCUGCUGUCUGG--AUUGGCCAAUGG------CAUCGACUGGCCCCAAUGGGCAGCCGCCACGCACCUGGCAAAGGUGUUUAAUGGGUAAU .-((.(((..(((.....(((....))))))..)))...))--((((.(((.(((------(...(.(((.(((....))))))).)))).((((((.....))))))....))).)))) ( -40.70) >DroSec_CAF1 41509 105 + 1 AACUUAUGUGUGCCAAUUGCC---UGCCGUCCGUUGCCUGCAUAUUGGCCAAUGGUUAUGGAUUUGACUGGCCUCAAUGGGCAGC---CACGCACCUGGCAA---------AUGGGUAAU .((((((...(((((..(((.---.((.((((((((..........(((((...((((......))))))))).)))))))).))---...)))..))))).---------))))))... ( -37.10) >DroSim_CAF1 42420 105 + 1 AACUUAUGUGUGCCAAUUGCC---UGCCGUCCGUUGCCUGCAUAUUGGCCAAUGGUUAUGGAUUUGACUGGCCUCAAUGGGCAGC---CACGCACCUGGCAA---------AUGGGUAAU .((((((...(((((..(((.---.((.((((((((..........(((((...((((......))))))))).)))))))).))---...)))..))))).---------))))))... ( -37.10) >DroYak_CAF1 41431 105 + 1 AACUUAUGUGUGCCAAUUGCC---UGCCGUCCGUUGCCUGCAUAUUGGCCAAUGGUUAUGGAUUUGACUGGCCUCAAUGGGCAGC---CACGCACCUGGCAA---------AUGGGUAAU .((((((...(((((..(((.---.((.((((((((..........(((((...((((......))))))))).)))))))).))---...)))..))))).---------))))))... ( -37.10) >consensus AACUUAUGUGUGCCAAUUGCC___UGCCGUCCGUUGCCUGCAUAUUGGCCAAUGGUUAUGGAUUUGACUGGCCUCAAUGGGCAGC___CACGCACCUGGCAA_________AUGGGUAAU .((((((.........(((((...(((.....((((((((......(((((...((((......)))))))))....))))))))......)))...))))).........))))))... (-28.35 = -29.43 + 1.08)

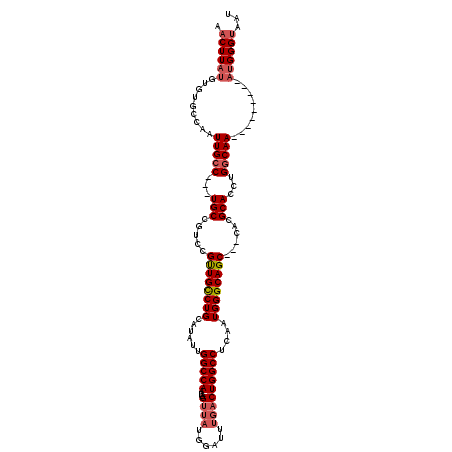

| Location | 18,521,122 – 18,521,227 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.31 |
| Mean single sequence MFE | -34.32 |
| Consensus MFE | -26.62 |
| Energy contribution | -27.22 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.544766 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18521122 105 - 22407834 AUUACCCAU---------UUGCCAGGUGCGUG---GCUGCCCAUUGAGGCCAGUCAAAUCCAUAACGAUUGGCCAAUAUGCAGGCAACGGACGGCA---GGCAAUUGGCACACAUAAGUU .........---------.((((((.(((.((---.(((.((.....((((((((...........))))))))........(....))).)))))---.))).)))))).......... ( -36.50) >DroPse_CAF1 50225 111 - 1 AUUACCCAUUAAACACCUUUGCCAGGUGCGUGGCGGCUGCCCAUUGGGGCCAGUCGAUG------CCAUUGGCCAAU--CCAGACAGCAGGCGGCAAUUGCCAAUUGGCCCACUGAAG-U .............(((((.....))))).(((((((((((((....))).))))))..(------(((...(((...--..........)))(((....)))...)))))))).....-. ( -37.52) >DroSec_CAF1 41509 105 - 1 AUUACCCAU---------UUGCCAGGUGCGUG---GCUGCCCAUUGAGGCCAGUCAAAUCCAUAACCAUUGGCCAAUAUGCAGGCAACGGACGGCA---GGCAAUUGGCACACAUAAGUU .........---------.((((((.(((.((---.(((.((.....(((((((.............)))))))........(....))).)))))---.))).)))))).......... ( -32.52) >DroSim_CAF1 42420 105 - 1 AUUACCCAU---------UUGCCAGGUGCGUG---GCUGCCCAUUGAGGCCAGUCAAAUCCAUAACCAUUGGCCAAUAUGCAGGCAACGGACGGCA---GGCAAUUGGCACACAUAAGUU .........---------.((((((.(((.((---.(((.((.....(((((((.............)))))))........(....))).)))))---.))).)))))).......... ( -32.52) >DroYak_CAF1 41431 105 - 1 AUUACCCAU---------UUGCCAGGUGCGUG---GCUGCCCAUUGAGGCCAGUCAAAUCCAUAACCAUUGGCCAAUAUGCAGGCAACGGACGGCA---GGCAAUUGGCACACAUAAGUU .........---------.((((((.(((.((---.(((.((.....(((((((.............)))))))........(....))).)))))---.))).)))))).......... ( -32.52) >consensus AUUACCCAU_________UUGCCAGGUGCGUG___GCUGCCCAUUGAGGCCAGUCAAAUCCAUAACCAUUGGCCAAUAUGCAGGCAACGGACGGCA___GGCAAUUGGCACACAUAAGUU ...................((((((.(((......((((.((.....(((((((.............)))))))........(....))).)))).....))).)))))).......... (-26.62 = -27.22 + 0.60)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:15:16 2006