
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,519,568 – 18,519,707 |
| Length | 139 |
| Max. P | 0.894235 |

| Location | 18,519,568 – 18,519,676 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 94.81 |
| Mean single sequence MFE | -27.62 |
| Consensus MFE | -25.29 |
| Energy contribution | -25.60 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.767975 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18519568 108 - 22407834 CAAAUGGGUUAGUGUGGCACUAGCCAGCAAUAAUAACAAAAAAAAAAAACACAAUUGCGGAAGCCAUCAAGUGUUUGGUAUGCGCAUUACUCAUGCGAACGCCCGUUU .(((((((((((((...)))))))).....................((((((.((.((....)).))...))))))(((...(((((.....)))))...)))))))) ( -28.00) >DroSec_CAF1 39957 104 - 1 CAAAUGGGUUAGUGUGGCACUAGCCAGCAAUAAUAACAGAA----AAGACACAAUUGCGGAAGCCAUCAAGUGUUUGGUAUGCGCAUUACUCAUGCGAUCGACCGUUU ......((((((((...))))))))(((.............----.((((((.((.((....)).))...))))))(((...(((((.....)))))....)))))). ( -28.50) >DroSim_CAF1 40861 104 - 1 CAAAUGGGUUAGUGUGGCACUAGCCAGCAAUAAUAACAGAA----AAAACACAAUUGCGGAAGCCAUCAAGUGUUUGGUAUGCGCAUUACUCAUGCGAACGCCCGUUU .((((((((..((.((((....))))))........((...----.((((((.((.((....)).))...))))))....))(((((.....)))))...)))))))) ( -28.50) >DroYak_CAF1 39876 103 - 1 CAAAUGGGUUAGUGUGGCACUAGCCUGCAAUAAUAACAGAA-----AAACACAAUUGCGGAAGCCAUCAAAUGUUUGGUAUGCGCAUUACUCAUGCGAACGCCCGUUU .((((((((..((((((((......))).............-----.........((((.(.((((.........)))).).)))).....)))))....)))))))) ( -25.50) >consensus CAAAUGGGUUAGUGUGGCACUAGCCAGCAAUAAUAACAGAA____AAAACACAAUUGCGGAAGCCAUCAAGUGUUUGGUAUGCGCAUUACUCAUGCGAACGCCCGUUU .(((((((((((((...)))))))).....................((((((.((.((....)).))...))))))(((...(((((.....)))))...)))))))) (-25.29 = -25.60 + 0.31)



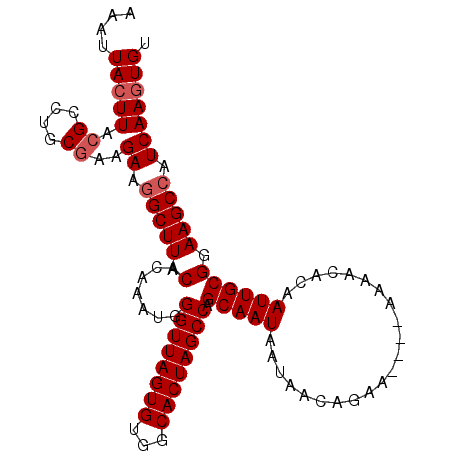
| Location | 18,519,602 – 18,519,707 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 93.69 |
| Mean single sequence MFE | -24.00 |
| Consensus MFE | -20.95 |
| Energy contribution | -21.70 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.894235 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18519602 105 - 22407834 AAAUUACUUACGCCUGCGAAGAAGGCUUACACAAAUGGGUUAGUGUGGCACUAGCCAGCAAUAAUAACAAAAAAAAAAAACACAAUUGCGGAAGCCAUCAAGUGU ....(((((.((....))..((.(((((.(.......((((((((...)))))))).(((((......................)))))).))))).))))))). ( -25.05) >DroSec_CAF1 39991 101 - 1 AAAUUACUUACGCCUGCGAAGAAAGCUUACACAAAUGGGUUAGUGUGGCACUAGCCAGCAAUAAUAACAGAA----AAGACACAAUUGCGGAAGCCAUCAAGUGU ....(((((.((....))..((..((((.(.......((((((((...)))))))).(((((..........----........)))))).))))..))))))). ( -21.17) >DroSim_CAF1 40895 101 - 1 AAAUUACUUACGCCUGCGAAGAAGGCUUACACAAAUGGGUUAGUGUGGCACUAGCCAGCAAUAAUAACAGAA----AAAACACAAUUGCGGAAGCCAUCAAGUGU ....(((((.((....))..((.(((((.(.......((((((((...)))))))).(((((..........----........)))))).))))).))))))). ( -25.27) >DroYak_CAF1 39910 100 - 1 AAAUUACUUGCGCCUGCCAAGAGGGCUUACACAAAUGGGUUAGUGUGGCACUAGCCUGCAAUAAUAACAGAA-----AAACACAAUUGCGGAAGCCAUCAAAUGU ..((((.((((((((.......))))..........(((((((((...))))))))))))))))).......-----..........((....)).......... ( -24.50) >consensus AAAUUACUUACGCCUGCGAAGAAGGCUUACACAAAUGGGUUAGUGUGGCACUAGCCAGCAAUAAUAACAGAA____AAAACACAAUUGCGGAAGCCAUCAAGUGU ....(((((.((....))..((.(((((.(.......((((((((...)))))))).(((((......................)))))).))))).))))))). (-20.95 = -21.70 + 0.75)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:15:12 2006