| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,519,256 – 18,519,391 |
| Length | 135 |
| Max. P | 0.993450 |

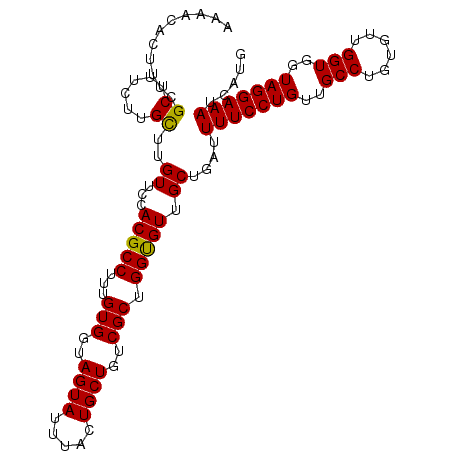
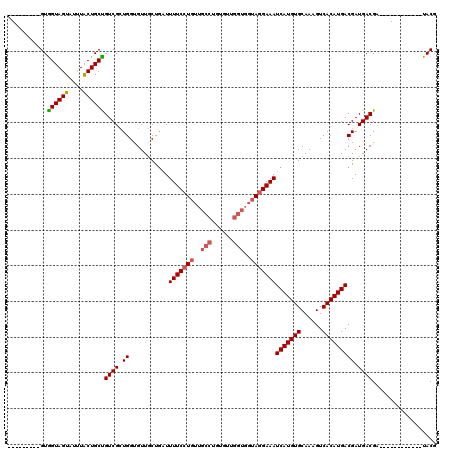
| Location | 18,519,256 – 18,519,363 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 98.13 |
| Mean single sequence MFE | -32.10 |
| Consensus MFE | -31.04 |
| Energy contribution | -30.60 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.955706 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18519256 107 + 22407834 AAAACACUUUCGCUUUCUUGUUUGUUCCACGCCUUUGUGGUAGUAUUUACUGCUGUCGCUGGCGUUGCUGAUUUUCCUGUUGCCUGUGUUGGUGGUAGGAAAUCAUG .(((((............)))))((...(((((...(((..((((.....))))..))).))))).))((((((.((((..(((......)))..)))))))))).. ( -33.00) >DroSec_CAF1 39645 107 + 1 AAAACACUUUCGCUUUCUUGCUUGUUCCACGCCUUUGUGGUAGUAUUUACUGCUGUCGCUGGUGUUGCUUAUUUUCCUGUUGCCUGUGUUGGUGGUAGGAAAUCAUG ...........((......))..((...(((((...(((..((((.....))))..))).))))).))....(((((((..(((......)))..)))))))..... ( -30.80) >DroSim_CAF1 40549 107 + 1 AAAACACUUUCGCUUUCUUGCUUGUUCCACGCCUUUGUGGUAGUAUUUACUGCUGUCGCUGGUGUUGCUGAUUUUCCUGUUGCCUGUGUUGGUGGUAGGAAAUCAUG ...........((......))..((...(((((...(((..((((.....))))..))).))))).))((((((.((((..(((......)))..)))))))))).. ( -32.50) >consensus AAAACACUUUCGCUUUCUUGCUUGUUCCACGCCUUUGUGGUAGUAUUUACUGCUGUCGCUGGUGUUGCUGAUUUUCCUGUUGCCUGUGUUGGUGGUAGGAAAUCAUG ...........((......))..((...(((((...(((..((((.....))))..))).))))).))....(((((((..(((......)))..)))))))..... (-31.04 = -30.60 + -0.44)

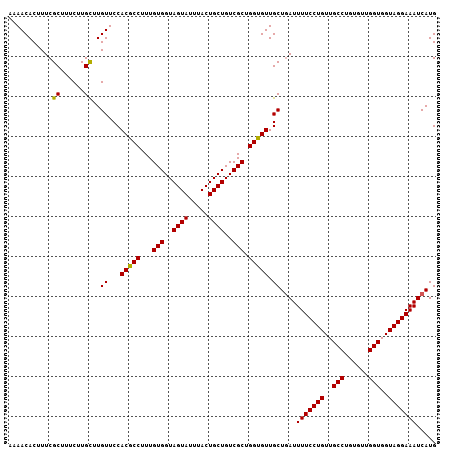
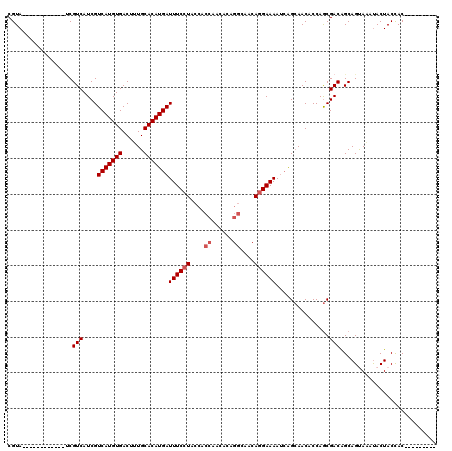
| Location | 18,519,292 – 18,519,391 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.61 |
| Mean single sequence MFE | -36.64 |
| Consensus MFE | -28.22 |
| Energy contribution | -28.66 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.77 |
| SVM decision value | 2.40 |
| SVM RNA-class probability | 0.993450 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18519292 99 + 22407834 ---------GUGGUAGUAUUUACUGCUGUCGCUGGCGUUGCUGAUUUUCCUGUUGCCUGUGUUGGUGGUAGGAAAUCAUGUGCAAAGUCACAUGACGAUGACGG------------UACG ---------..((((((....))))))((.((((.(((((.....(((((((..(((......)))..)))))))(((((((......)))))))))))).)))------------))). ( -35.60) >DroPse_CAF1 48192 114 + 1 UGUGAAGUAGUAGUAGUGGCUGCUGCUGUCGUUGGUGUUCCCGCUUUUCGU------CGUGUUGGUGGUAGGAAAUCAUGUGCAAAGUCACAUGACGAUGACGAUGACGAUGACUUUACG .(((((((((((((((...))))))))(((((((...((((..((..(((.------.....))).))..)))).(((((((......))))))))))))))..........))))))). ( -38.80) >DroSec_CAF1 39681 99 + 1 ---------GUGGUAGUAUUUACUGCUGUCGCUGGUGUUGCUUAUUUUCCUGUUGCCUGUGUUGGUGGUAGGAAAUCAUGUGCAAAGUCACAUGACGAUGACGA------------UACG ---------(((..((((.....))))..)))..((((((.(((((((((((..(((......)))..)))))).(((((((......))))))).))))))))------------))). ( -36.20) >DroSim_CAF1 40585 99 + 1 ---------GUGGUAGUAUUUACUGCUGUCGCUGGUGUUGCUGAUUUUCCUGUUGCCUGUGUUGGUGGUAGGAAAUCAUGUGCAAAGUCACAUGACGAUGACGA------------UACG ---------(((..((((.....))))..)))..((((((.....(((((((..(((......)))..)))))))(((((((......)))))))......)))------------))). ( -34.80) >DroYak_CAF1 39601 98 + 1 ---------GUGGUAGUAU-UGCUGCCGUCGCUGGUGUUGCUGAUUUUCCUGUUGCCUGUGUUGGUGGUAGGAAAUCAUGUGCAAAGUCACAUGACGAUGACGC------------UACG ---------..(((((...-..)))))..((.(((((((..((..(((((((..(((......)))..)))))))(((((((......)))))))))..)))))------------)))) ( -37.80) >consensus _________GUGGUAGUAUUUACUGCUGUCGCUGGUGUUGCUGAUUUUCCUGUUGCCUGUGUUGGUGGUAGGAAAUCAUGUGCAAAGUCACAUGACGAUGACGA____________UACG ...........((((((....))))))((((.((...........(((((((..(((......)))..)))))))(((((((......))))))))).)))).................. (-28.22 = -28.66 + 0.44)

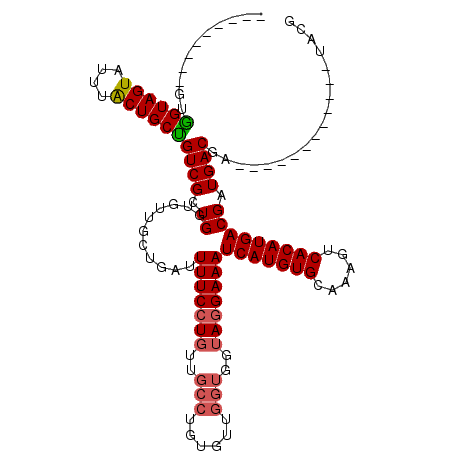
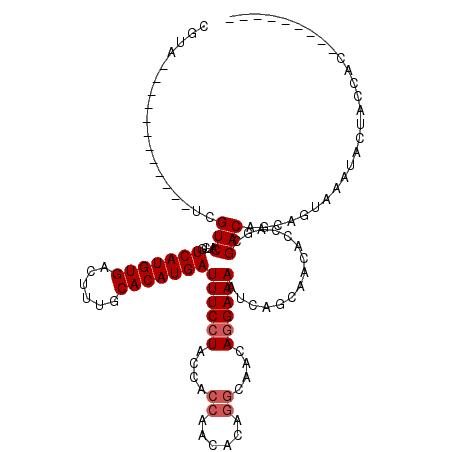
| Location | 18,519,292 – 18,519,391 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.61 |
| Mean single sequence MFE | -20.58 |
| Consensus MFE | -16.12 |
| Energy contribution | -16.72 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.567718 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18519292 99 - 22407834 CGUA------------CCGUCAUCGUCAUGUGACUUUGCACAUGAUUUCCUACCACCAACACAGGCAACAGGAAAAUCAGCAACGCCAGCGACAGCAGUAAAUACUACCAC--------- ..((------------(.((..((((((((((......)))))).((((((....((......))....)))))).............))))..)).)))...........--------- ( -19.90) >DroPse_CAF1 48192 114 - 1 CGUAAAGUCAUCGUCAUCGUCAUCGUCAUGUGACUUUGCACAUGAUUUCCUACCACCAACACG------ACGAAAAGCGGGAACACCAACGACAGCAGCAGCCACUACUACUACUUCACA .((.((((....(((.(((((...((((((((......))))))))................)------)))).....((.....))...))).((....))..........)))).)). ( -21.71) >DroSec_CAF1 39681 99 - 1 CGUA------------UCGUCAUCGUCAUGUGACUUUGCACAUGAUUUCCUACCACCAACACAGGCAACAGGAAAAUAAGCAACACCAGCGACAGCAGUAAAUACUACCAC--------- ....------------..(((....(((((((......)))))))((((((....((......))....))))))....((.......)))))..................--------- ( -19.30) >DroSim_CAF1 40585 99 - 1 CGUA------------UCGUCAUCGUCAUGUGACUUUGCACAUGAUUUCCUACCACCAACACAGGCAACAGGAAAAUCAGCAACACCAGCGACAGCAGUAAAUACUACCAC--------- ....------------..(((....(((((((......)))))))((((((....((......))....))))))....((.......)))))..................--------- ( -19.30) >DroYak_CAF1 39601 98 - 1 CGUA------------GCGUCAUCGUCAUGUGACUUUGCACAUGAUUUCCUACCACCAACACAGGCAACAGGAAAAUCAGCAACACCAGCGACGGCAGCA-AUACUACCAC--------- ....------------(((((.((((((((((......)))))).((((((....((......))....)))))).............)))).))).)).-..........--------- ( -22.70) >consensus CGUA____________UCGUCAUCGUCAUGUGACUUUGCACAUGAUUUCCUACCACCAACACAGGCAACAGGAAAAUCAGCAACACCAGCGACAGCAGUAAAUACUACCAC_________ ..................(((....(((((((......)))))))((((((....((......))....))))))...............)))........................... (-16.12 = -16.72 + 0.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:15:10 2006