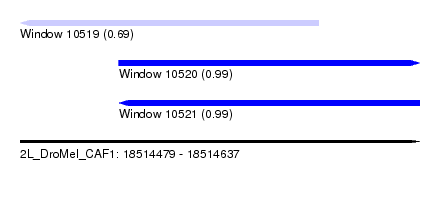
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,514,479 – 18,514,637 |
| Length | 158 |
| Max. P | 0.989944 |

| Location | 18,514,479 – 18,514,597 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.24 |
| Mean single sequence MFE | -30.77 |
| Consensus MFE | -11.70 |
| Energy contribution | -12.50 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.38 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.689635 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18514479 118 - 22407834 UUACAAACUAUAAUUUACUUUAAUGGCCAUCUGUAUUUUAUU-UGUGGCUCGUUGUAGUAGGCCGGCCAUUUGGCCAAAGCU-UCAAUGUUCGGAGUUGAUGCAUAAAUAAUGGGUUAAA ...............((((.((((((((((..(((...))).-.))))).))))).))))((((((((....))))...((.-(((((.......))))).))..........))))... ( -31.70) >DroPse_CAF1 41935 119 - 1 GUACAAGCCAUAAUUUGGCUUAAUGGCCAUCUGCGUUUUGCCGCGUGUGUACACGCAGUAGGCCUCCU-CGGAGCCCCAGCUCUUAGUGCUUCAGGCUAAUGCAUAAAUAAACCCAUAAA ....((((((.....)))))).((((.....((((((..((((((((....))))).(.((((...((-.(((((....))))).)).))))).))).)))))).........))))... ( -38.54) >DroSec_CAF1 34913 118 - 1 UUACAAACUAUAAUUUGCUUUAAUGGCCAUCCGUAUUUUAUU-UGUGUCUCGUUGUAGUAGGCCGGCCAUUUGGCCAAAGCU-UCACUGGUCGGAGUUGAUGCAUAAAUAAUGUGAUAAA (((((...........(((.....)))..........(((((-(((((.((.........((((((....))))))..((((-((.......)))))))).))))))))))))))).... ( -26.90) >DroSim_CAF1 33466 118 - 1 UUACAAACUAUAAUUUGCUUUAAUGGCCAUCUGUAUUUUAUU-UGUGGCUCGUUGUAGUAGGCCGGCCAUUUGGCCAAAGCU-UCACUGGUCGGAGUUGAUGCAUAAAUAAUGUGAUAAA .....((((....((((((.((((((((((..(((...))).-.))))).))))).))))))(((((((...(((....)))-....)))))))))))...((((.....))))...... ( -31.20) >DroYak_CAF1 34849 99 - 1 UUACAAACUAUAAUUUGCU-------------------UAUU-UGUGACUCGUUGUAGUAGGCCGGUCAUUCGGCAAAAGCU-UUAAUGGUCGGAGCUAGUGCAUAAAAAAUGGUUUUAA ....(((((((....((((-------------------((.(-(.((((.(((((.(((..(((((....)))))....)))-.))))))))).)).))).)))......)))))))... ( -25.50) >consensus UUACAAACUAUAAUUUGCUUUAAUGGCCAUCUGUAUUUUAUU_UGUGGCUCGUUGUAGUAGGCCGGCCAUUUGGCCAAAGCU_UCAAUGGUCGGAGUUGAUGCAUAAAUAAUGGGAUAAA ......................................................(((.(((.(((((((((.(((....)))...)))))))))..))).)))................. (-11.70 = -12.50 + 0.80)

| Location | 18,514,518 – 18,514,637 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.53 |
| Mean single sequence MFE | -32.02 |
| Consensus MFE | -16.88 |
| Energy contribution | -18.08 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.53 |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.986849 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18514518 119 + 22407834 CUUUGGCCAAAUGGCCGGCCUACUACAACGAGCCACA-AAUAAAAUACAGAUGGCCAUUAAAGUAAAUUAUAGUUUGUAAGACUAUACAUACAAUUUGACUGUUUUGUCGGGCCAAGAUC .(((((((.(((((((((((.........).)))...-......((....)))))))))...(((...(((((((.....)))))))..)))....((((......)))))))))))... ( -31.30) >DroPse_CAF1 41975 107 + 1 CUGGGGCUCCG-AGGAGGCCUACUGCGUGUACACACGCGGCAAAACGCAGAUGGCCAUUAAGCCAAAUUAUGGCUUGUACGG----------AAUUCGGCUCUUAUGCUGGGCCGA--GG (((((.(((..-..))).))..(((((((....))))))).......)))....((..(((((((.....)))))))...))----------..((((((((.......)))))))--). ( -43.60) >DroSec_CAF1 34952 119 + 1 CUUUGGCCAAAUGGCCGGCCUACUACAACGAGACACA-AAUAAAAUACGGAUGGCCAUUAAAGCAAAUUAUAGUUUGUAAGACUAUACAUCCCAUUUGACUGUUUUGUCGGGCCAAGAGC ((((((((....))))(((((......((((((((((-(((.......(((((((.......))....(((((((.....)))))))))))).)))))..)))))))).))))).)))). ( -33.80) >DroSim_CAF1 33505 119 + 1 CUUUGGCCAAAUGGCCGGCCUACUACAACGAGCCACA-AAUAAAAUACAGAUGGCCAUUAAAGCAAAUUAUAGUUUGUAAGACUAUACAUACCAUUUGACUGUUUUGUCGGGCCAAGAGC .(((((((...((((((...........)).))))..-.((((((((((((((((.......))....(((((((.....))))))).....))))))..))))))))..)))))))... ( -30.90) >DroYak_CAF1 34888 100 + 1 CUUUUGCCGAAUGACCGGCCUACUACAACGAGUCACA-AAUA-------------------AGCAAAUUAUAGUUUGUAAGACUAUACAUAUAAUUUGACUGUUUUGUCUGCUGAAGAGC ((((.((.((.((((((...........)).))))((-((..-------------------(((((((((((...((((......))))))))))))).))..)))))).)).))))... ( -20.50) >consensus CUUUGGCCAAAUGGCCGGCCUACUACAACGAGACACA_AAUAAAAUACAGAUGGCCAUUAAAGCAAAUUAUAGUUUGUAAGACUAUACAUACAAUUUGACUGUUUUGUCGGGCCAAGAGC ....((((........))))...............................(((((............(((((((.....))))))).........((((......)))))))))..... (-16.88 = -18.08 + 1.20)

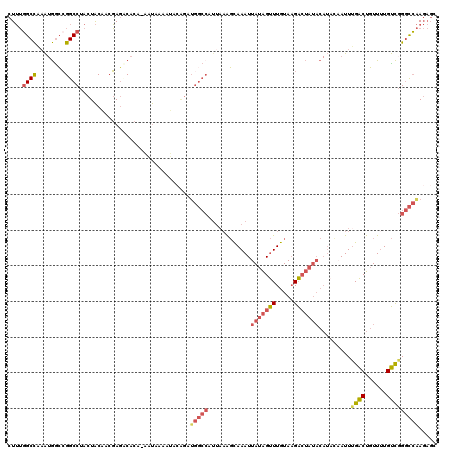
| Location | 18,514,518 – 18,514,637 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.53 |
| Mean single sequence MFE | -33.64 |
| Consensus MFE | -14.42 |
| Energy contribution | -15.42 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.43 |
| SVM decision value | 2.19 |
| SVM RNA-class probability | 0.989944 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
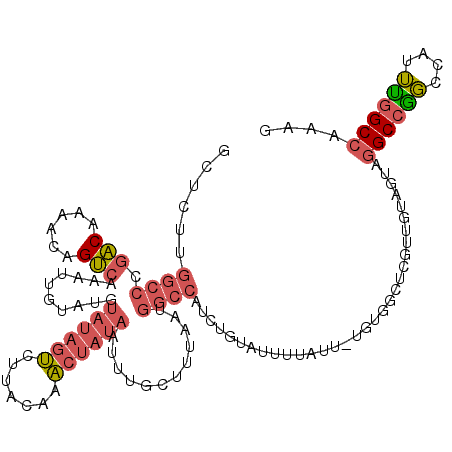
>2L_DroMel_CAF1 18514518 119 - 22407834 GAUCUUGGCCCGACAAAACAGUCAAAUUGUAUGUAUAGUCUUACAAACUAUAAUUUACUUUAAUGGCCAUCUGUAUUUUAUU-UGUGGCUCGUUGUAGUAGGCCGGCCAUUUGGCCAAAG ......((((.(((......)))((((((((((((......))))...))))))))(((.((((((((((..(((...))).-.))))).))))).))).))))((((....)))).... ( -36.00) >DroPse_CAF1 41975 107 - 1 CC--UCGGCCCAGCAUAAGAGCCGAAUU----------CCGUACAAGCCAUAAUUUGGCUUAAUGGCCAUCUGCGUUUUGCCGCGUGUGUACACGCAGUAGGCCUCCU-CGGAGCCCCAG ..--(((((.(.......).))))).((----------(((...((((((.....))))))...((((..((((((..((((....).))).))))))..))))....-)))))...... ( -36.80) >DroSec_CAF1 34952 119 - 1 GCUCUUGGCCCGACAAAACAGUCAAAUGGGAUGUAUAGUCUUACAAACUAUAAUUUGCUUUAAUGGCCAUCCGUAUUUUAUU-UGUGUCUCGUUGUAGUAGGCCGGCCAUUUGGCCAAAG ......((((..((((.((((..((((((((((((((((.......))))))....(((.....)))))))).)))))...)-)))......))))....))))((((....)))).... ( -32.20) >DroSim_CAF1 33505 119 - 1 GCUCUUGGCCCGACAAAACAGUCAAAUGGUAUGUAUAGUCUUACAAACUAUAAUUUGCUUUAAUGGCCAUCUGUAUUUUAUU-UGUGGCUCGUUGUAGUAGGCCGGCCAUUUGGCCAAAG ......((((.(((......)))..(((((.((((......)))).)))))....((((.((((((((((..(((...))).-.))))).))))).))))))))((((....)))).... ( -38.80) >DroYak_CAF1 34888 100 - 1 GCUCUUCAGCAGACAAAACAGUCAAAUUAUAUGUAUAGUCUUACAAACUAUAAUUUGCU-------------------UAUU-UGUGACUCGUUGUAGUAGGCCGGUCAUUCGGCAAAAG .((((.(((((((((((..((.(((((((((((((......))))...)))))))))))-------------------..))-)))..)).)))).)).))(((((....)))))..... ( -24.40) >consensus GCUCUUGGCCCGACAAAACAGUCAAAUUGUAUGUAUAGUCUUACAAACUAUAAUUUGCUUUAAUGGCCAUCUGUAUUUUAUU_UGUGGCUCGUUGUAGUAGGCCGGCCAUUUGGCCAAAG ......((((.(((......)))..........((((((.......))))))............))))................................((((((....)))))).... (-14.42 = -15.42 + 1.00)

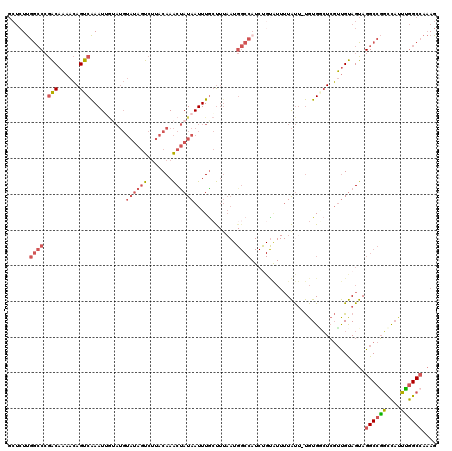
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:15:03 2006