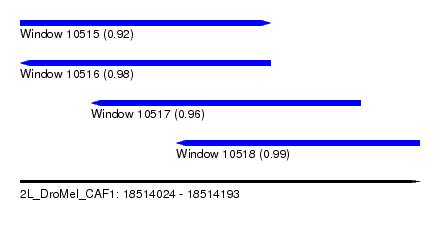
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,514,024 – 18,514,193 |
| Length | 169 |
| Max. P | 0.994147 |

| Location | 18,514,024 – 18,514,130 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.01 |
| Mean single sequence MFE | -24.39 |
| Consensus MFE | -16.82 |
| Energy contribution | -17.22 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918466 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18514024 106 + 22407834 AUAAAAUAGUUAGUCGUUUAAUUUAAAUC----C---------UCAUUA-AAAGCCACAACAAAAGGAAGUUGCCGAAGGAGCAGCCGUUGGCUUCAUCAUGAGGAAGCCAGAAAUGCUG ..........(((.(((((........((----(---------((((..-.(((((((..(....)...((((((....).))))).).))))))....)))))))......)))))))) ( -25.04) >DroPse_CAF1 41193 119 + 1 AUAAAUUAUUAAGUCGUUUAAUUUAAACCAAUCCCCAACCAUCCCAUAAAACAAACAC-ACAAAAGAGUGUUGCCGAGGGAGCAACCGACUGCUUCGAAAUGACUAAGCAACAAUUGCUU ...........((((((((..................................(((((-........)))))..(((((.(((....).)).)))))))))))))(((((.....))))) ( -22.00) >DroSec_CAF1 34458 106 + 1 AUAAAUUAGUAAGUCGUUUAAUUUAAAUC----C---------UAAUAA-AAAGCCACAACAAAAGGAAGUUGCCGCAGGAGCAGCCGUUGGCUUCAUCAUGACGAAGCCAGAAAUGCUG .((((((((........))))))))..((----(---------(.....-...((..((((........))))..)))))).((((..((((((((........))))))))....)))) ( -25.40) >DroSim_CAF1 33006 106 + 1 AUAAAUUAGUAAGUCGUUUAAUUUAAAUC----C---------UAAUAA-AAAGCCACAACAAAAGGAAGUUGCCGCAGGAGCAGCCGUUGGCUUCAUCAUGACGAAGCCAGAAAUGCUG .((((((((........))))))))..((----(---------(.....-...((..((((........))))..)))))).((((..((((((((........))))))))....)))) ( -25.40) >DroYak_CAF1 34436 94 + 1 AUAAAUUAGUAAGUCGUUUAAUUUAAUUC----C---------UCAUAA-A------------AAGGAAGUUGACGCAGGAGUAGCCGCUGGCUUCAUCAUGACGAAGCCAGAAAUGCUG .((((((((........)))))))).(((----(---------(.....-.------------.)))))......(((((.....)).((((((((........))))))))...))).. ( -24.10) >consensus AUAAAUUAGUAAGUCGUUUAAUUUAAAUC____C_________UCAUAA_AAAGCCACAACAAAAGGAAGUUGCCGCAGGAGCAGCCGUUGGCUUCAUCAUGACGAAGCCAGAAAUGCUG .((((((((........)))))))).......................................................((((....((((((((........))))))))...)))). (-16.82 = -17.22 + 0.40)

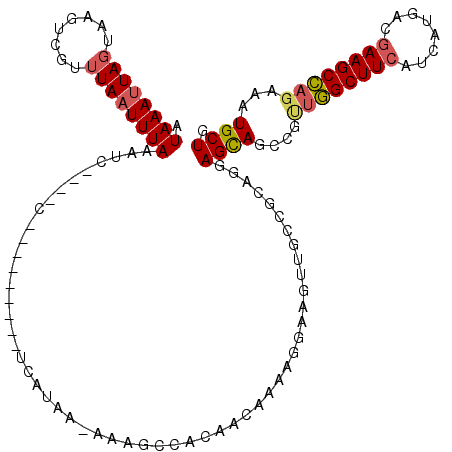
| Location | 18,514,024 – 18,514,130 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.01 |
| Mean single sequence MFE | -28.18 |
| Consensus MFE | -14.16 |
| Energy contribution | -15.80 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.50 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.975819 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18514024 106 - 22407834 CAGCAUUUCUGGCUUCCUCAUGAUGAAGCCAACGGCUGCUCCUUCGGCAACUUCCUUUUGUUGUGGCUUU-UAAUGA---------G----GAUUUAAAUUAAACGACUAACUAUUUUAU .(((.......)))(((((((...((((((..(((........)))(((((........)))))))))))-..))))---------)----))........................... ( -25.80) >DroPse_CAF1 41193 119 - 1 AAGCAAUUGUUGCUUAGUCAUUUCGAAGCAGUCGGUUGCUCCCUCGGCAACACUCUUUUGU-GUGUUUGUUUUAUGGGAUGGUUGGGGAUUGGUUUAAAUUAAACGACUUAAUAAUUUAU ..(((((((((((((.(......).)))))).)))))))((((..((((((((.(....).-))).)))))....)))).(((((..((((......))))...)))))........... ( -30.80) >DroSec_CAF1 34458 106 - 1 CAGCAUUUCUGGCUUCGUCAUGAUGAAGCCAACGGCUGCUCCUGCGGCAACUUCCUUUUGUUGUGGCUUU-UUAUUA---------G----GAUUUAAAUUAAACGACUUACUAAUUUAU ((((.....(((((((((....)))))))))...)))).((((((.(((((........))))).))...-.....)---------)----))..(((((((..........))))))). ( -29.70) >DroSim_CAF1 33006 106 - 1 CAGCAUUUCUGGCUUCGUCAUGAUGAAGCCAACGGCUGCUCCUGCGGCAACUUCCUUUUGUUGUGGCUUU-UUAUUA---------G----GAUUUAAAUUAAACGACUUACUAAUUUAU ((((.....(((((((((....)))))))))...)))).((((((.(((((........))))).))...-.....)---------)----))..(((((((..........))))))). ( -29.70) >DroYak_CAF1 34436 94 - 1 CAGCAUUUCUGGCUUCGUCAUGAUGAAGCCAGCGGCUACUCCUGCGUCAACUUCCUU------------U-UUAUGA---------G----GAAUUAAAUUAAACGACUUACUAAUUUAU .(((....((((((((((....))))))))))..)))..............((((((------------.-....))---------)----))).(((((((..........))))))). ( -24.90) >consensus CAGCAUUUCUGGCUUCGUCAUGAUGAAGCCAACGGCUGCUCCUGCGGCAACUUCCUUUUGUUGUGGCUUU_UUAUGA_________G____GAUUUAAAUUAAACGACUUACUAAUUUAU ((((.....(((((((((....)))))))))...))))........(((((........)))))...............................(((((((..........))))))). (-14.16 = -15.80 + 1.64)

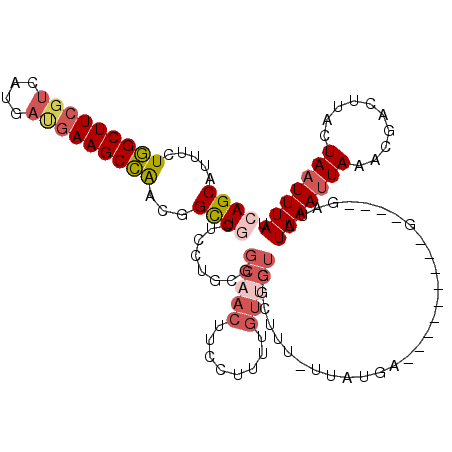
| Location | 18,514,054 – 18,514,168 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.43 |
| Mean single sequence MFE | -26.40 |
| Consensus MFE | -15.32 |
| Energy contribution | -16.96 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.961021 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18514054 114 - 22407834 AAUUAAGAAUUUUCA--ACUCACCAUAAAAUUAAACUAAUCAGCAUUUCUGGCUUCCUCAUGAUGAAGCCAACGGCUGCUCCUUCGGCAACUUCCUUUUGUUGUGGCUUU-UAAUGA--- .(((((((.......--.......................((((.....(((((((........)))))))...))))......(.(((((........))))).).)))-))))..--- ( -22.60) >DroPse_CAF1 41233 119 - 1 AUUUAGGAACUGCCAAAACUCACCAUAAAAUUAAACUAAAAAGCAAUUGUUGCUUAGUCAUUUCGAAGCAGUCGGUUGCUCCCUCGGCAACACUCUUUUGU-GUGUUUGUUUUAUGGGAU .....((.....)).....((.(((((((((.((((.....((((((((((((((.(......).)))))).)))))))).........((((......))-))))))))))))))))). ( -30.80) >DroSec_CAF1 34488 114 - 1 AAUUAAGAAUUUUCA--ACUCACCAUAAAAUUAAACUAAUCAGCAUUUCUGGCUUCGUCAUGAUGAAGCCAACGGCUGCUCCUGCGGCAACUUCCUUUUGUUGUGGCUUU-UUAUUA--- ...((((((......--.......................((((.....(((((((((....)))))))))...)))).....((.(((((........))))).)))))-)))...--- ( -28.40) >DroSim_CAF1 33036 114 - 1 AAUUAAGAAUUUUCU--ACUCACCAUAAAAUUAAACUAAUCAGCAUUUCUGGCUUCGUCAUGAUGAAGCCAACGGCUGCUCCUGCGGCAACUUCCUUUUGUUGUGGCUUU-UUAUUA--- ...((((((......--.......................((((.....(((((((((....)))))))))...)))).....((.(((((........))))).)))))-)))...--- ( -28.40) >DroYak_CAF1 34466 102 - 1 AAUUAAAAACUUUCA--ACUCACCAUAAAAUUAAACUAAUCAGCAUUUCUGGCUUCGUCAUGAUGAAGCCAGCGGCUACUCCUGCGUCAACUUCCUU------------U-UUAUGA--- ...............--......(((((((............(((...((((((((((....)))))))))).((.....)))))...........)------------)-))))).--- ( -21.80) >consensus AAUUAAGAAUUUUCA__ACUCACCAUAAAAUUAAACUAAUCAGCAUUUCUGGCUUCGUCAUGAUGAAGCCAACGGCUGCUCCUGCGGCAACUUCCUUUUGUUGUGGCUUU_UUAUGA___ ....................(((.((((((..........((((.....(((((((((....)))))))))...)))).......(....)....)))))).)))............... (-15.32 = -16.96 + 1.64)


| Location | 18,514,090 – 18,514,193 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.23 |
| Mean single sequence MFE | -18.61 |
| Consensus MFE | -13.46 |
| Energy contribution | -13.70 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.72 |
| SVM decision value | 2.45 |
| SVM RNA-class probability | 0.994147 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18514090 103 - 22407834 UCGGA------CAAAUCACAC-AUUAAAUAC--------UAAUUAAGAAUUUUCA--ACUCACCAUAAAAUUAAACUAAUCAGCAUUUCUGGCUUCCUCAUGAUGAAGCCAACGGCUGCU .....------..........-.........--------................--.......................((((.....(((((((........)))))))...)))).. ( -14.60) >DroPse_CAF1 41272 97 - 1 UCAU--------------CUA-AAUAAAAAC--------UAUUUAGGAACUGCCAAAACUCACCAUAAAAUUAAACUAAAAAGCAAUUGUUGCUUAGUCAUUUCGAAGCAGUCGGUUGCU ...(--------------(((-((((.....--------))))))))..................................((((((((((((((.(......).)))))).)))))))) ( -19.00) >DroSec_CAF1 34524 109 - 1 UCGGACUUUAACAAAUCACAC-AUUAAAUAU--------UAAUUAAGAAUUUUCA--ACUCACCAUAAAAUUAAACUAAUCAGCAUUUCUGGCUUCGUCAUGAUGAAGCCAACGGCUGCU .....................-.........--------................--.......................((((.....(((((((((....)))))))))...)))).. ( -17.80) >DroSim_CAF1 33072 109 - 1 UCGGACUUUAACAAAUCACAC-AUUAAAUAC--------UAAUUAAGAAUUUUCU--ACUCACCAUAAAAUUAAACUAAUCAGCAUUUCUGGCUUCGUCAUGAUGAAGCCAACGGCUGCU ..((.................-((((.....--------))))..(((....)))--.....))................((((.....(((((((((....)))))))))...)))).. ( -17.90) >DroYak_CAF1 34490 107 - 1 -----------AAAAUGACACCAACAAAUGCUAAUGGCUUAAUUAAAAACUUUCA--ACUCACCAUAAAAUUAAACUAAUCAGCAUUUCUGGCUUCGUCAUGAUGAAGCCAGCGGCUACU -----------.........((...(((((((..(((.((((((...........--...........)))))).)))...)))))))((((((((((....)))))))))).))..... ( -23.75) >consensus UCGGA______CAAAUCACAC_AUUAAAUAC________UAAUUAAGAAUUUUCA__ACUCACCAUAAAAUUAAACUAAUCAGCAUUUCUGGCUUCGUCAUGAUGAAGCCAACGGCUGCU ................................................................................((((.....(((((((((....)))))))))...)))).. (-13.46 = -13.70 + 0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:15:00 2006