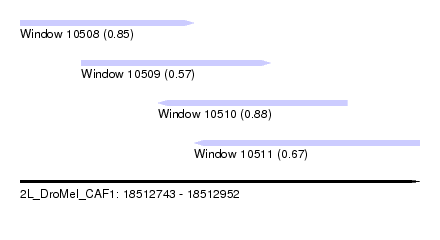
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,512,743 – 18,512,952 |
| Length | 209 |
| Max. P | 0.875079 |

| Location | 18,512,743 – 18,512,834 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 98.90 |
| Mean single sequence MFE | -34.73 |
| Consensus MFE | -33.90 |
| Energy contribution | -34.15 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.845854 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18512743 91 + 22407834 CCAGAUCGAGACUGGGUCUGGGAAUGUCUACGGUCUGGGGUAAUGGGGUCUUGGGUGCUUACAUCACAGCAGGAGAUCCCAGACUCGAGAC (((((((.......)))))))...........((((.((((..(((((((((...((((........)))).))))))))).)))).)))) ( -35.50) >DroSec_CAF1 33186 91 + 1 CCAGAUCGAGACUGGGUCUGGGAAUGUCUAUGGUCUGGGGUAAUGGGGUCUUGGGUGCUUACAUCACAGCAGGAGAUCCCAGACUCGAGAC (((((((.......)))))))...........((((.((((..(((((((((...((((........)))).))))))))).)))).)))) ( -35.50) >DroSim_CAF1 31739 91 + 1 CCAGAUCGAGACUGGGUCUGGGAAUGUCUAUGGUCUGGGGUAAUGGGGUCUUGGGUGCUUACAUCACAGCAGGAGAUCCCAGACUCGAGAC (((((((.......)))))))...........((((.((((..(((((((((...((((........)))).))))))))).)))).)))) ( -35.50) >DroYak_CAF1 33162 91 + 1 CCAGAUCGAGACUGGGUCUUGGAAUGUCUAUGGUCUGGGGUAAUGGGGUCUUGGGUGCUUACAUCACAGCAGGAGAUCCCAGACUCGAGAC .((..(((((((...)))))))..))......((((.((((..(((((((((...((((........)))).))))))))).)))).)))) ( -32.40) >consensus CCAGAUCGAGACUGGGUCUGGGAAUGUCUAUGGUCUGGGGUAAUGGGGUCUUGGGUGCUUACAUCACAGCAGGAGAUCCCAGACUCGAGAC (((((((.......)))))))...........((((.((((..(((((((((...((((........)))).))))))))).)))).)))) (-33.90 = -34.15 + 0.25)

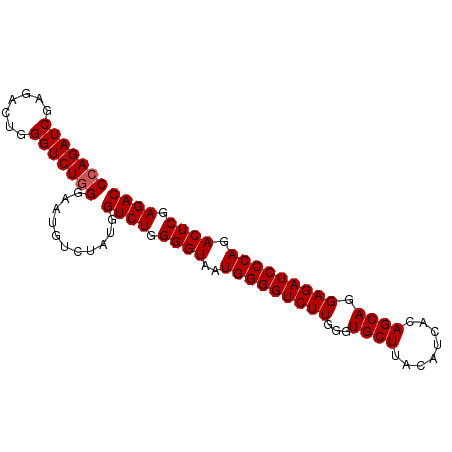
| Location | 18,512,775 – 18,512,874 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.15 |
| Mean single sequence MFE | -34.16 |
| Consensus MFE | -25.35 |
| Energy contribution | -25.67 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.570273 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18512775 99 + 22407834 GUCUGGGGUAAUGGGGUCUUGGGUGCUUACAUCACAGCAGGAGA----UC-----------------CCAGACUCGAGACUCGUCCCACAAUAAGAUGAAACUUGUUUCCUCUCGACUCU ((((.((((..(((((((((...((((........)))).))))----))-----------------))).)))).))))(((......((((((......))))))......))).... ( -31.10) >DroPse_CAF1 39790 120 + 1 GUAAUGGGUAAUGGGGUUUUGGGCGCUUACAUCACAGCAGGAGGCAGACCCGACAGCAAGAUGGAGUCUCGUCUCGAGACUCGUCUCACAAUGAGAUGAAACUUGUUUCCUCUCGACUCU .....((((...((((..((((((((((.(.........).)))).).))))).((((((...((((((((...))))))))(((((.....)))))....)))))).))))...)))). ( -39.00) >DroSec_CAF1 33218 99 + 1 GUCUGGGGUAAUGGGGUCUUGGGUGCUUACAUCACAGCAGGAGA----UC-----------------CCAGACUCGAGACUCGUCCCACAAUGAGAUGAAACUUGUUUCCUCUCGACUCU ((((.((((..(((((((((...((((........)))).))))----))-----------------))).)))).))))...........(((((.((((....)))).)))))..... ( -34.80) >DroSim_CAF1 31771 99 + 1 GUCUGGGGUAAUGGGGUCUUGGGUGCUUACAUCACAGCAGGAGA----UC-----------------CCAGACUCGAGACUCGUCCCACAAUGAGAUGAAACUUGUUUCCUCUCGACUCU ((((.((((..(((((((((...((((........)))).))))----))-----------------))).)))).))))...........(((((.((((....)))).)))))..... ( -34.80) >DroYak_CAF1 33194 99 + 1 GUCUGGGGUAAUGGGGUCUUGGGUGCUUACAUCACAGCAGGAGA----UC-----------------CCAGACUCGAGACUCGUCCCACAAUAAGAAGAAACUUGUUUCCUCUCGACUCU ((((.((((..(((((((((...((((........)))).))))----))-----------------))).)))).))))(((......((((((......))))))......))).... ( -31.10) >consensus GUCUGGGGUAAUGGGGUCUUGGGUGCUUACAUCACAGCAGGAGA____UC_________________CCAGACUCGAGACUCGUCCCACAAUGAGAUGAAACUUGUUUCCUCUCGACUCU (((..(((.....((((((((((((((........))).((..........................))..)))))))))))..))).....((((.((((....)))).)))))))... (-25.35 = -25.67 + 0.32)

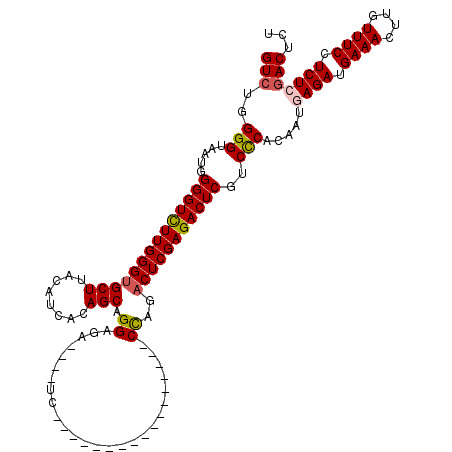
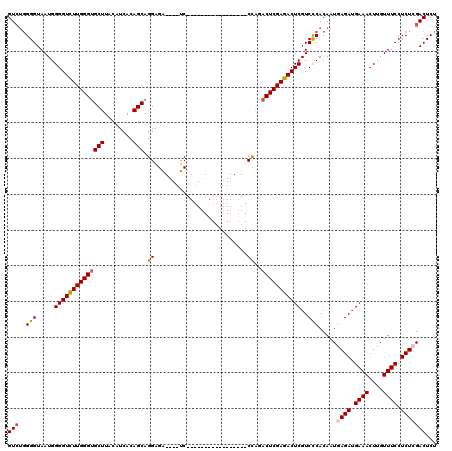
| Location | 18,512,815 – 18,512,914 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.99 |
| Mean single sequence MFE | -32.18 |
| Consensus MFE | -23.62 |
| Energy contribution | -24.58 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.875079 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
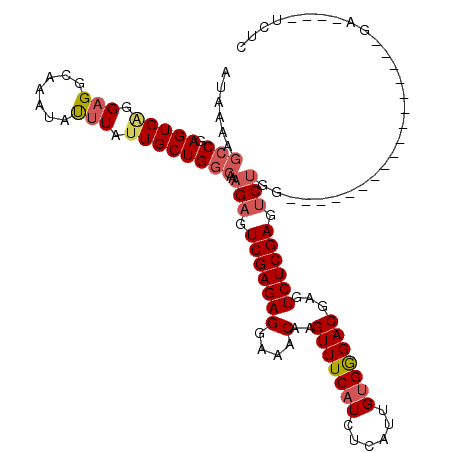
>2L_DroMel_CAF1 18512815 99 - 22407834 AUAAAAGCCGAGUGAGGAGGCAAAUACUUAUUGCUGGCAAAGAGUCGAGAGGAAACAAGUUUCAUCUUAUUGUGGGACGAGUCUCGAGUCUGG-----------------GA----UCUC ......(((.((..(.(((.......))).)..)))))..(((.(((((((....)..((..(((......)))..))...)))))).)))..-----------------..----.... ( -31.50) >DroPse_CAF1 39830 117 - 1 ---GACCGCGAGUGGGGAGUCAAAUAUUUAUUGCUGGCAAAGAGUCGAGAGGAAACAAGUUUCAUCUCAUUGUGAGACGAGUCUCGAGACGAGACUCCAUCUUGCUGUCGGGUCUGCCUC ---(((.(((((...((((((.........(((((.((((.(((..(((((....)...))))..))).)))).)).)))(((....)))..))))))..))))).)))(((....))). ( -42.50) >DroSec_CAF1 33258 99 - 1 AUAAAAGCCGAGUGAGGAGGCAAAUAUUUGUUGCUGGCAAAGAGUCGAGAGGAAACAAGUUUCAUCUCAUUGUGGGACGAGUCUCGAGUCUGG-----------------GA----UCUC ......(((.((..(.(((.......))).)..)))))..(((.(((((((....)..((..(((......)))..))...)))))).)))..-----------------..----.... ( -30.60) >DroSim_CAF1 31811 99 - 1 AUAAAAGCCGAGUGAGGAGGCAAAUAUUUGUUGCUGGCAAAGAGUCGAGAGGAAACAAGUUUCAUCUCAUUGUGGGACGAGUCUCGAGUCUGG-----------------GA----UCUC ......(((.((..(.(((.......))).)..)))))..(((.(((((((....)..((..(((......)))..))...)))))).)))..-----------------..----.... ( -30.60) >DroYak_CAF1 33234 99 - 1 AUAAAAGCCGAGUGAGGUGGCAAAUAUUUACUGCUGGCAAAGAGUCGAGAGGAAACAAGUUUCUUCUUAUUGUGGGACGAGUCUCGAGUCUGG-----------------GA----UCUC ......((((((((((..........))))))..))))..(((.(((((((((((....))))))))).....((((....)))))).)))..-----------------..----.... ( -25.70) >consensus AUAAAAGCCGAGUGAGGAGGCAAAUAUUUAUUGCUGGCAAAGAGUCGAGAGGAAACAAGUUUCAUCUCAUUGUGGGACGAGUCUCGAGUCUGG_________________GA____UCUC ......(((.(((((.(((.......))).))))))))..(((.(((((((....)..(((((((......)))))))...)))))).)))............................. (-23.62 = -24.58 + 0.96)

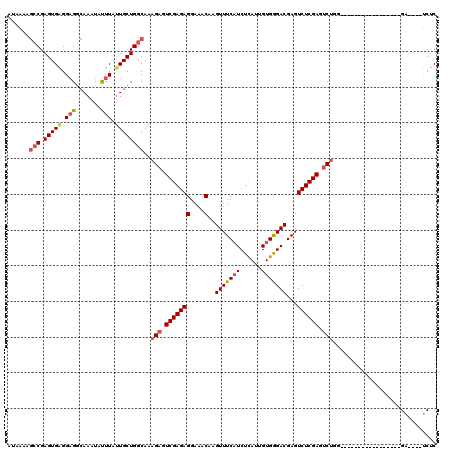
| Location | 18,512,834 – 18,512,952 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.97 |
| Mean single sequence MFE | -25.44 |
| Consensus MFE | -18.74 |
| Energy contribution | -19.42 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.668050 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
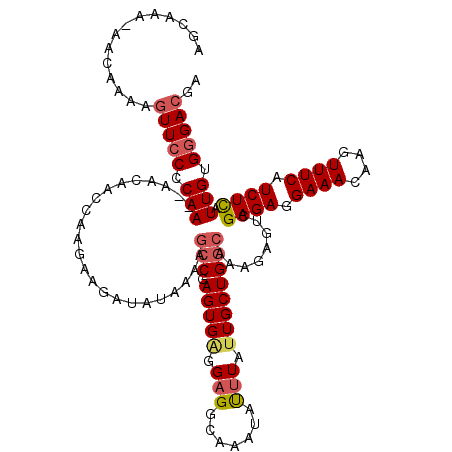
>2L_DroMel_CAF1 18512834 118 - 22407834 AGCAAAAAACAAAAGUUCCCCAA--AACAACCAAGAAGAUAUAAAAGCCGAGUGAGGAGGCAAAUACUUAUUGCUGGCAAAGAGUCGAGAGGAAACAAGUUUCAUCUUAUUGUGGGACGA ..............(((((.(((--.....................(((.((..(.(((.......))).)..)))))........((((.((((....)))).)))).))).))))).. ( -25.10) >DroPse_CAF1 39870 104 - 1 AUAAAA-UACAGAGAUUCCACAA--AAAAA-------------GACCGCGAGUGGGGAGUCAAAUAUUUAUUGCUGGCAAAGAGUCGAGAGGAAACAAGUUUCAUCUCAUUGUGAGACGA ......-......((((((.(..--.....-------------).(((....))))))))).........(((((.((((.(((..(((((....)...))))..))).)))).)).))) ( -22.60) >DroSec_CAF1 33277 117 - 1 AGCAAA-AACAAAAGUUCCCCAA--AACAACCAAGAAGAUAUAAAAGCCGAGUGAGGAGGCAAAUAUUUGUUGCUGGCAAAGAGUCGAGAGGAAACAAGUUUCAUCUCAUUGUGGGACGA ......-.......(((((.(((--.....................(((.((..(.(((.......))).)..)))))........((((.((((....)))).)))).))).))))).. ( -26.80) >DroSim_CAF1 31830 117 - 1 AGCAAA-AACAAAAGUUCCCCAA--AACAACCAAGAAGAUAUAAAAGCCGAGUGAGGAGGCAAAUAUUUGUUGCUGGCAAAGAGUCGAGAGGAAACAAGUUUCAUCUCAUUGUGGGACGA ......-.......(((((.(((--.....................(((.((..(.(((.......))).)..)))))........((((.((((....)))).)))).))).))))).. ( -26.80) >DroYak_CAF1 33253 119 - 1 AGCAAA-AACAAAGGUUCCCCAAAAAACAACCAAGAAGAUAUAAAAGCCGAGUGAGGUGGCAAAUAUUUACUGCUGGCAAAGAGUCGAGAGGAAACAAGUUUCUUCUUAUUGUGGGACGA ......-.......(((((.(((.......(((((.((((((....(((......))).....)))))).))..))).........(((((((((....))))))))).))).))))).. ( -25.90) >consensus AGCAAA_AACAAAAGUUCCCCAA__AACAACCAAGAAGAUAUAAAAGCCGAGUGAGGAGGCAAAUAUUUAUUGCUGGCAAAGAGUCGAGAGGAAACAAGUUUCAUCUCAUUGUGGGACGA ..............(((((.(((.......................(((.(((((.(((.......))).))))))))........((((.((((....)))).)))).))).))))).. (-18.74 = -19.42 + 0.68)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:14:54 2006