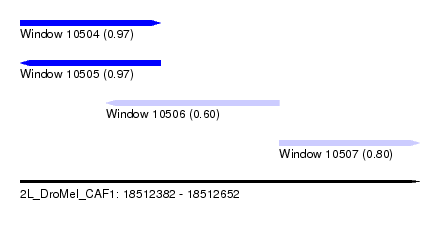
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,512,382 – 18,512,652 |
| Length | 270 |
| Max. P | 0.974136 |

| Location | 18,512,382 – 18,512,477 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 90.35 |
| Mean single sequence MFE | -27.05 |
| Consensus MFE | -22.58 |
| Energy contribution | -23.20 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.972830 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
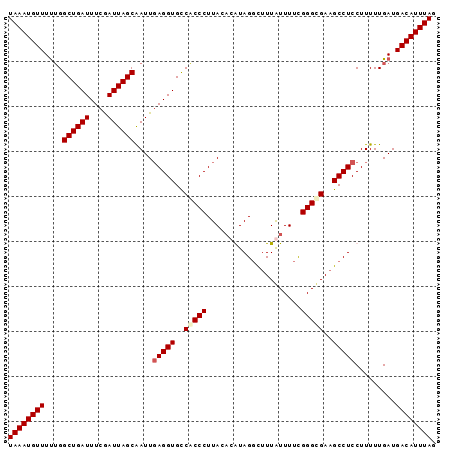
>2L_DroMel_CAF1 18512382 95 + 22407834 UAAAUGUUUUUGGCUGAUUUCGAUUAGCAAUUGAGGUGCCGCCCUUACACAUAGGCAUUAUUUUCGGGCGAAGCCUCCUUUUUGAUGACAUUUAG ((((((((.((.((((((....))))))....(((((..(((((.....................)))))..)))))......)).)))))))). ( -27.80) >DroSec_CAF1 32825 95 + 1 UAAAUGUUUUUGGCUGAUUUCGAUUAGCGAUUGAGGUGCCACCCUUACACGUAGGUUUUGUUUUCGGGCGAAGCCUCCUUUUUGAUGACAUUUAG ((((((((..((((..(((((((((...))))))))))))).........(.((((((((((....)))))))))).)........)))))))). ( -25.30) >DroSim_CAF1 31378 95 + 1 UAAAUGUUUUUGGCUGAUUUCGAUUAGCGAUUGAGGUGCCACCCUUAAACGUAGGUUUUGUUUUCGGGCGAAGCCUCCUUUUUGAUGACAUUUAG ((((((((.((.((((((....))))))....(((((..(.(((..(((((.......)))))..))).)..)))))......)).)))))))). ( -27.70) >DroYak_CAF1 32810 93 + 1 UAAAUGUUUCAGGCUGAUUUCAAUUAGCAAUUCAGGUGUCACCCUUAC--AUAGUCUUUAUUUUUGGGUGAAGCCUCCUUUUUGAUGACAUUUAG ((((((((((((((((((....)))))).....((((.((((((....--...............)))))).)))).....)))).)))))))). ( -27.41) >consensus UAAAUGUUUUUGGCUGAUUUCGAUUAGCAAUUGAGGUGCCACCCUUACACAUAGGCUUUAUUUUCGGGCGAAGCCUCCUUUUUGAUGACAUUUAG ((((((((....((((((....))))))....(((((..(((((.....................)))))..))))).........)))))))). (-22.58 = -23.20 + 0.62)

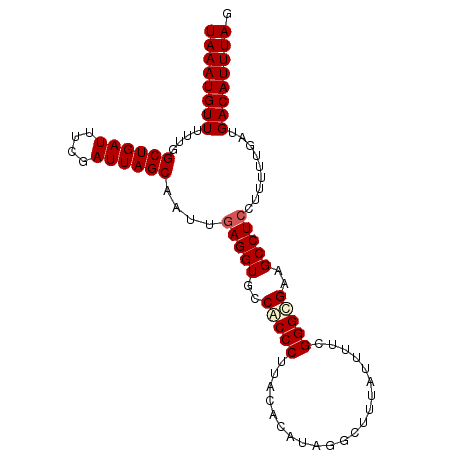
| Location | 18,512,382 – 18,512,477 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 90.35 |
| Mean single sequence MFE | -20.55 |
| Consensus MFE | -18.46 |
| Energy contribution | -18.15 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.974136 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18512382 95 - 22407834 CUAAAUGUCAUCAAAAAGGAGGCUUCGCCCGAAAAUAAUGCCUAUGUGUAAGGGCGGCACCUCAAUUGCUAAUCGAAAUCAGCCAAAAACAUUUA .(((((((..........((((..((((((....(((.......)))....))))))..))))....(((.((....)).))).....))))))) ( -21.50) >DroSec_CAF1 32825 95 - 1 CUAAAUGUCAUCAAAAAGGAGGCUUCGCCCGAAAACAAAACCUACGUGUAAGGGUGGCACCUCAAUCGCUAAUCGAAAUCAGCCAAAAACAUUUA .(((((((..........((((..((((((....((.........))....))))))..))))....(((.((....)).))).....))))))) ( -20.40) >DroSim_CAF1 31378 95 - 1 CUAAAUGUCAUCAAAAAGGAGGCUUCGCCCGAAAACAAAACCUACGUUUAAGGGUGGCACCUCAAUCGCUAAUCGAAAUCAGCCAAAAACAUUUA .(((((((..........((((..((((((..((((.........))))..))))))..))))....(((.((....)).))).....))))))) ( -22.00) >DroYak_CAF1 32810 93 - 1 CUAAAUGUCAUCAAAAAGGAGGCUUCACCCAAAAAUAAAGACUAU--GUAAGGGUGACACCUGAAUUGCUAAUUGAAAUCAGCCUGAAACAUUUA .(((((((..(((......(((..((((((....(((.....)))--....))))))..))).....(((.((....)).))).))).))))))) ( -18.30) >consensus CUAAAUGUCAUCAAAAAGGAGGCUUCGCCCGAAAACAAAACCUACGUGUAAGGGUGGCACCUCAAUCGCUAAUCGAAAUCAGCCAAAAACAUUUA .(((((((..........((((..((((((.....................))))))..))))....(((.((....)).))).....))))))) (-18.46 = -18.15 + -0.31)

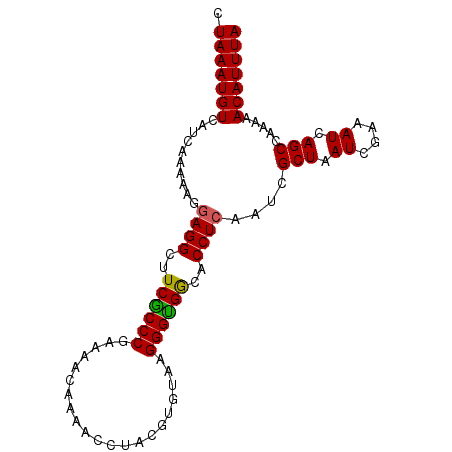
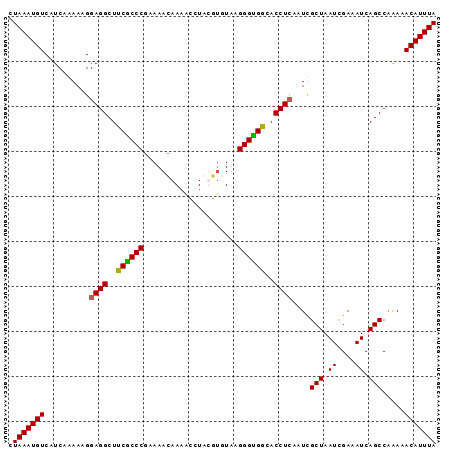
| Location | 18,512,440 – 18,512,557 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.09 |
| Mean single sequence MFE | -34.02 |
| Consensus MFE | -25.54 |
| Energy contribution | -25.34 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.604097 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18512440 117 - 22407834 UCGUUUAGUAGUCGGGUGCUAAAAUGCCAGGCAAACACCGCCGACAGGUGGCGCAUGUCCGACAGCCAUAAAUUGGACAGCUAAAUGUCAUCAAAAAGGAGGCUUCGCCCGAAAAUA--- ...........(((((((.......(((.(((.......)))((((..((((...(((((((..........)))))))))))..))))...........)))..))))))).....--- ( -37.30) >DroPse_CAF1 39412 114 - 1 UCGAUUAGUGUCGAGGCGCUAAAAUGCCAAGCAAACAGGGACGACAGGUGGCGCAUGUCUGUUGGCCAUAAAUUGGAUGGCUAAAUGUCAUCAAAAAG-AGGCU-----CAAAACCAUCA ..(((((((((....)))))))...(((...(((....((.((((((..(.....)..)))))).)).....)))((((((.....))))))......-.))).-----........)). ( -32.00) >DroSec_CAF1 32883 117 - 1 UCGAUUAGUAGCCGGGUGCUAAAAUGCCAGGCAAACACCGCCGACAGGUGGCGCAUGUCCGUCAGCCAUAAAUUGGACAGCUAAAUGUCAUCAAAAAGGAGGCUUCGCCCGAAAACA--- ............((((((.......(((.(((.......)))((((..((((...((((((............))))))))))..))))...........)))..))))))......--- ( -35.30) >DroSim_CAF1 31436 117 - 1 UCGAUUAGUAGCCGGGUGCUAAAAUGCCAGGCAAACACCGCCGACAGGUGGCGCAUGUCCGUCAGCCAUAAAUUGGACAGCUAAAUGUCAUCAAAAAGGAGGCUUCGCCCGAAAACA--- ............((((((.......(((.(((.......)))((((..((((...((((((............))))))))))..))))...........)))..))))))......--- ( -35.30) >DroYak_CAF1 32866 117 - 1 UCGAUUAGUAGCCGGGUGCUAAAAUGCCAGUCAAACACCGCCGACAGGUGGCGCAUGUCCGACAGCCAUAAAUUGGACAGCUAAAUGUCAUCAAAAAGGAGGCUUCACCCAAAAAUA--- .............(((((.......(((.........(((((....))))).((.(((((((..........)))))))))...................)))..))))).......--- ( -30.20) >consensus UCGAUUAGUAGCCGGGUGCUAAAAUGCCAGGCAAACACCGCCGACAGGUGGCGCAUGUCCGUCAGCCAUAAAUUGGACAGCUAAAUGUCAUCAAAAAGGAGGCUUCGCCCGAAAACA___ ....((((((......))))))...(((.(((.......)))((((..((((...((((((............))))))))))..))))...........)))................. (-25.54 = -25.34 + -0.20)

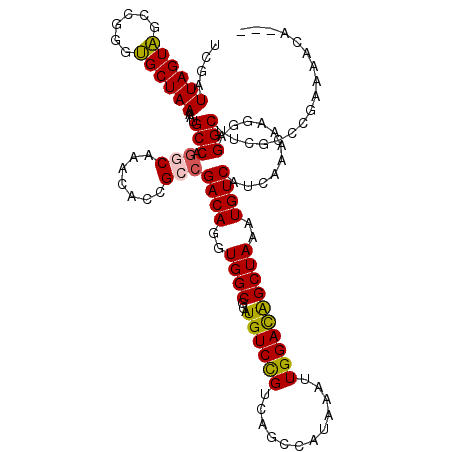
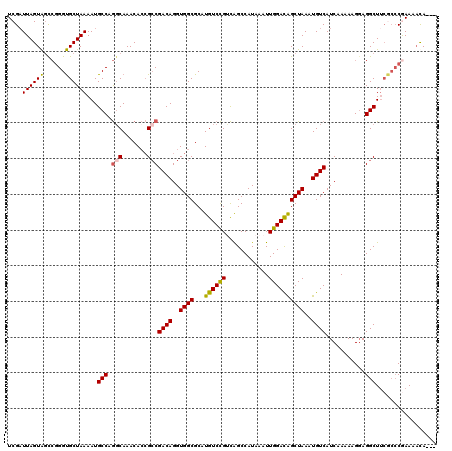
| Location | 18,512,557 – 18,512,652 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.84 |
| Mean single sequence MFE | -31.38 |
| Consensus MFE | -22.54 |
| Energy contribution | -23.70 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.802483 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18512557 95 + 22407834 AAUGUUUGGCCAAACAAUCAACGUGGCCAA-----------------------GUCAGCUUUUGUGUCUGGGCAUCGCGG-GGAUGCCCUAUCACUGAUGGUUUGCACUCGAUUUCUAA- .....(((((((...........)))))))-----------------------((.((((...(((...(((((((....-.)))))))...)))....)))).)).............- ( -30.50) >DroPse_CAF1 39526 119 + 1 AAUGUUUGGCCAGAUAAUUAACUUGGCCGACAAGAGAUGUACCCAAGAGAUGGGUCUGGUC-UGGGUCUGGUCGUGGCGGGGGAUGCCCUCUCGCUGAUAAGUUGCAUUCGAUUUCUAAU (((((..((((((((......((((.....))))((((..(((((.....)))))...)))-)..))))))))..((((((((.....))))))))........)))))........... ( -37.70) >DroSec_CAF1 33000 95 + 1 AAUGUUUGGCUAAACAAUCAACUUGGCCAA-----------------------GUCAGCUUUGGUGUCUGGGCAUCGCGG-GGAUGCCCUAUCACUGAUGGUUUGCAUUCGAUUUCUAA- .....((((((((.........))))))))-----------------------((.(((((..(((...(((((((....-.)))))))...)))..).)))).)).............- ( -31.80) >DroSim_CAF1 31553 95 + 1 AAUGUUUGGCCAAACAAUCAACUUGGCCAA-----------------------AUCAGCUUUUGUGUCUGGGCAUCGCGG-GGAUGCCCUAUCACUGAUGGUUUGCAUUCGAUUUCUAA- ...((((((((((.........))))))))-----------------------)).((((...(((...(((((((....-.)))))))...)))....))))................- ( -30.60) >DroYak_CAF1 32983 94 + 1 AAUGUUUGGCCAAACAAUCAACUUCGCCAA-----------------------GUCAGCUU-UGUGUCUGGGCAUCGCGG-GGAUGCCCUAUAACUGAUGGCUUGCAUUCGAUUUCUAA- (((((..(((((............(((.((-----------------------(....)))-.)))...(((((((....-.))))))).........))))).)))))..........- ( -26.30) >consensus AAUGUUUGGCCAAACAAUCAACUUGGCCAA_______________________GUCAGCUUUUGUGUCUGGGCAUCGCGG_GGAUGCCCUAUCACUGAUGGUUUGCAUUCGAUUUCUAA_ .....((((((((.........)))))))).......................((.((((...(((...(((((((......)))))))...)))....)))).)).............. (-22.54 = -23.70 + 1.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:14:50 2006