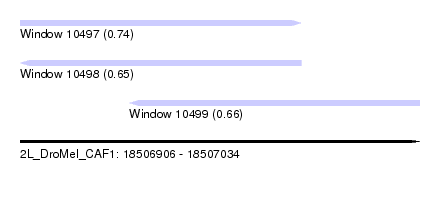
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,506,906 – 18,507,034 |
| Length | 128 |
| Max. P | 0.742159 |

| Location | 18,506,906 – 18,506,996 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 92.27 |
| Mean single sequence MFE | -18.77 |
| Consensus MFE | -15.50 |
| Energy contribution | -16.88 |
| Covariance contribution | 1.38 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.742159 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
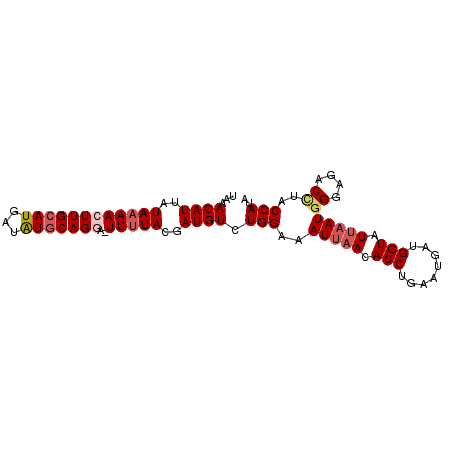
>2L_DroMel_CAF1 18506906 90 + 22407834 UAAACAUUAUAAAACUUGCAUGAUGUUCAGGAAUUGUACGAUGUCUGGAAAUUAACACCUUAAUGAUGGUAU-AAUGGGAGACAUACCAUA ....((((((..........((((.(((((..(((....)))..))))).))))..(((........)))))-))))(....)........ ( -15.20) >DroSec_CAF1 27285 90 + 1 UAAACAUUAUAAAACUUGCAUGAUAUGCAGCA-UUUUACGAUGUCUGGAAAUUAACACCUGAAUGAUGGUAUUAAUGGGAGACCUACCAUA ...((((..(((((.((((((...))))))..-)))))..)))).(((..(((((.(((........))).)))))((....))..))).. ( -20.80) >DroSim_CAF1 26479 90 + 1 UAAACAUUAUAAAACUUGCAUGAUAUGCAGGA-UUUUACGAUGUCUGGAAAUGAACACCUUAAUGAUGGUAUUAAUGGGAGACCUACCAUA ...((((..((((((((((((...))))))).-)))))..)))).(((........(((........)))......((....))..))).. ( -22.50) >DroYak_CAF1 27355 90 + 1 AAAACAUAAUAAAACUUGCAUGAUGUGCAGAA-UUUUACGAUGUCUGGAAAUUAAGACCUGAAUGAUGGUAUUAAUGGGAGACUUACCAUA ...((((..(((((.((((((...))))))..-)))))..)))).(((..(((((.(((........))).)))))((....))..))).. ( -16.60) >consensus UAAACAUUAUAAAACUUGCAUGAUAUGCAGGA_UUUUACGAUGUCUGGAAAUUAACACCUGAAUGAUGGUAUUAAUGGGAGACCUACCAUA ...((((..((((((((((((...)))))))..)))))..)))).(((..(((((.(((........))).)))))((....))..))).. (-15.50 = -16.88 + 1.38)

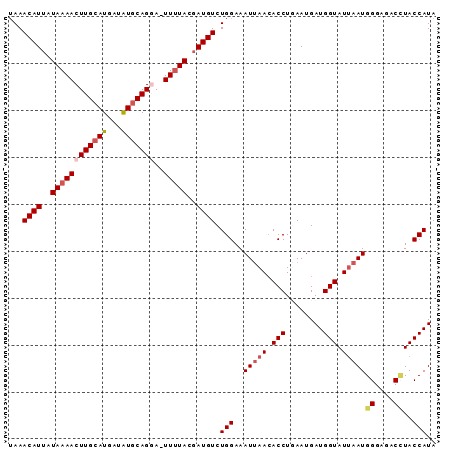
| Location | 18,506,906 – 18,506,996 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 92.27 |
| Mean single sequence MFE | -18.55 |
| Consensus MFE | -13.52 |
| Energy contribution | -14.78 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.646632 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18506906 90 - 22407834 UAUGGUAUGUCUCCCAUU-AUACCAUCAUUAAGGUGUUAAUUUCCAGACAUCGUACAAUUCCUGAACAUCAUGCAAGUUUUAUAAUGUUUA .((((((((........)-)))))))......((((((........))))))..........(((((((.(((.......))).))))))) ( -15.60) >DroSec_CAF1 27285 90 - 1 UAUGGUAGGUCUCCCAUUAAUACCAUCAUUCAGGUGUUAAUUUCCAGACAUCGUAAAA-UGCUGCAUAUCAUGCAAGUUUUAUAAUGUUUA ...((..((....))(((((((((........)))))))))..))((((((..(((((-(..(((((...))))).))))))..)))))). ( -23.30) >DroSim_CAF1 26479 90 - 1 UAUGGUAGGUCUCCCAUUAAUACCAUCAUUAAGGUGUUCAUUUCCAGACAUCGUAAAA-UCCUGCAUAUCAUGCAAGUUUUAUAAUGUUUA .((((........)))).((((((........)))))).......((((((..(((((-(..(((((...))))).))))))..)))))). ( -18.80) >DroYak_CAF1 27355 90 - 1 UAUGGUAAGUCUCCCAUUAAUACCAUCAUUCAGGUCUUAAUUUCCAGACAUCGUAAAA-UUCUGCACAUCAUGCAAGUUUUAUUAUGUUUU ..(((........)))((((.(((........))).)))).....((((((.((((((-((.((((.....)))))))))))).)))))). ( -16.50) >consensus UAUGGUAGGUCUCCCAUUAAUACCAUCAUUAAGGUGUUAAUUUCCAGACAUCGUAAAA_UCCUGCACAUCAUGCAAGUUUUAUAAUGUUUA ..(((........)))((((((((........)))))))).....((((((..(((((....((((.....))))..)))))..)))))). (-13.52 = -14.78 + 1.25)

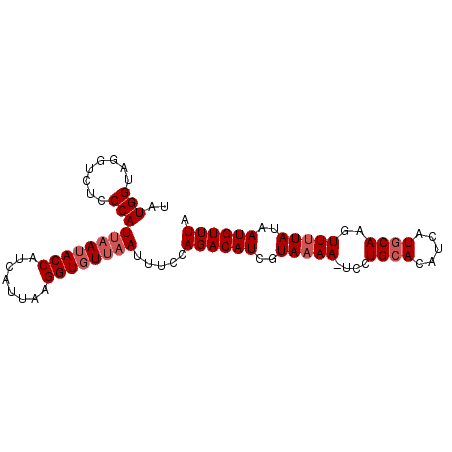
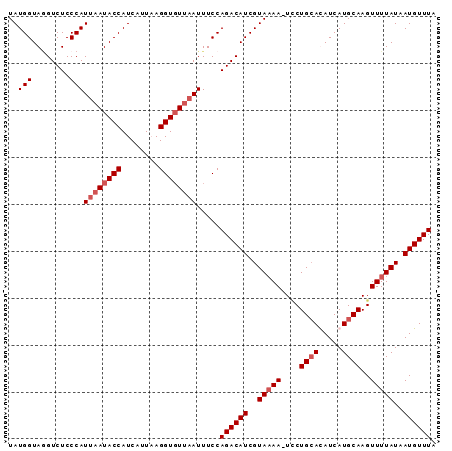
| Location | 18,506,941 – 18,507,034 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.93 |
| Mean single sequence MFE | -24.36 |
| Consensus MFE | -9.42 |
| Energy contribution | -11.14 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.39 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.664630 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
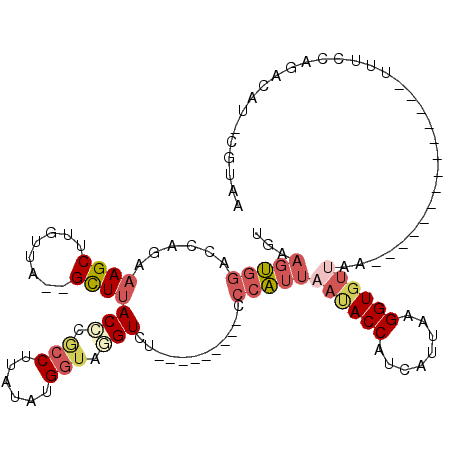
>2L_DroMel_CAF1 18506941 93 - 22407834 UGAAGUGGACCAGAAAGCUUGUUA--GCUUACCCGCCUUAUAUGGUAUGUCU---------CCCAUU-AUACCAUCAUUAAGGUGUUAA--------------UUUCCAGACAU-CGUAC .....((((.....(((((....)--))))...(((((((.((((((((...---------.....)-)))))))...)))))))....--------------..)))).....-..... ( -24.70) >DroPse_CAF1 32456 117 - 1 UAGCCAUAGCC--AUAGCUUUAUACGGUUUACGCCUCAUAUAUG-UAUGUAUGUUAGUGUGGCUUUUACCACCAUAAUAAAGGUGCCUAGACUCUGGCCUGUUUGGCUAGACAUUCGAGU .((((..((((--.((......)).)))).((((..((((((..-..))))))...)))))))).....((((........))))....((.(((((((.....)))))))...)).... ( -29.40) >DroSec_CAF1 27319 94 - 1 CGAAGUGGACCAGAAAGCUUGUUA--GCUUACCCGCCUUAUAUGGUAGGUCU---------CCCAUUAAUACCAUCAUUCAGGUGUUAA--------------UUUCCAGACAU-CGUAA (((.(((((((...(((((....)--))))....(((......))).)))))---------...(((((((((........))))))))--------------)......)).)-))... ( -24.60) >DroSim_CAF1 26513 94 - 1 CGAAGUGGACCAGAAAGCUUGUUA--GCUUACCCGCCUUGUAUGGUAGGUCU---------CCCAUUAAUACCAUCAUUAAGGUGUUCA--------------UUUCCAGACAU-CGUAA .(((((((((....(((((....)--))))....((((((.((((((.....---------........))))))...)))))))))))--------------)))).......-..... ( -25.12) >DroYak_CAF1 27389 94 - 1 UGAAGUGGACCAGAAAGCUUGUCA--GCUUACCCACCUUAUAUGGUAAGUCU---------CCCAUUAAUACCAUCAUUCAGGUCUUAA--------------UUUCCAGACAU-CGUAA (((.((((....(.(((((....)--)))).))))).)))((((((..((((---------...(((((.(((........))).))))--------------)....))))))-)))). ( -18.00) >consensus UGAAGUGGACCAGAAAGCUUGUUA__GCUUACCCGCCUUAUAUGGUAGGUCU_________CCCAUUAAUACCAUCAUUAAGGUGUUAA______________UUUCCAGACAU_CGUAA ...(((((......((((........))))(((.(((......))).)))............)))))((((((........))))))................................. ( -9.42 = -11.14 + 1.72)

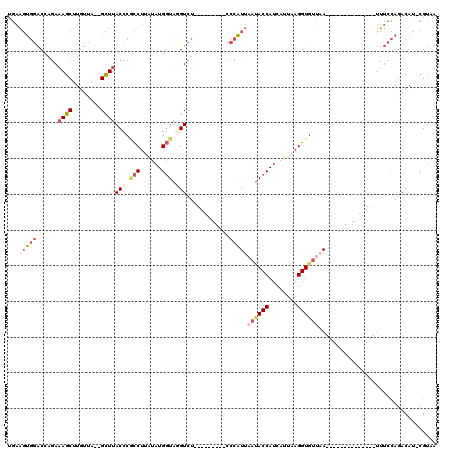
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:14:43 2006