
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,496,581 – 18,496,672 |
| Length | 91 |
| Max. P | 0.926411 |

| Location | 18,496,581 – 18,496,672 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 92.32 |
| Mean single sequence MFE | -24.52 |
| Consensus MFE | -18.11 |
| Energy contribution | -18.75 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.926411 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18496581 91 + 22407834 CUGUGUCGCAGCUUCUGGUUACCUGGUUUUUAUUAAAAAAGUUAGUUUUUU-CGAACUCACCUUUCAUGGCUGCGACAGUUUGCAAUUGCAA ...((((((((((...(((...((..(((((.....)))))..))......-.(....))))......))))))))))..((((....)))) ( -23.50) >DroSec_CAF1 17081 89 + 1 CUGUGUCGCAGCUUCUGGUUACCUGCUUUUUAUUAAA-AAGUUGGUU-UUU-CGAACUCACCUUUCAUGGCUGCGACAGUUUGCAAUUGCAA ...((((((((((...(((.(((.((((((.....))-)))).))).-...-.......)))......))))))))))..((((....)))) ( -27.30) >DroSim_CAF1 16379 90 + 1 CUGUGUCGCAGCUUCUGGUUACCUGCUUUUUAUUAAA-AAUUUGGUUUUUU-CGAACUCACCUUUCAUGGCUGCGACAGUUUGCAAUUGCAA ...((((((((((...(((.(((....((((....))-))...))).....-.(....))))......))))))))))..((((....)))) ( -22.00) >DroEre_CAF1 17573 90 + 1 CUGUGUCGCAGCUUCUUGUUGGCUGCUUUUUACUAAA-AAGUUGAUUUUUUU-GAACUCACCUUUCAUGGCUGCGACAGUUUGCAAUUGCAA ...((((((((((...((..((..((((((.....))-))))((((((....-))).)))))...)).))))))))))..((((....)))) ( -24.30) >DroYak_CAF1 17084 91 + 1 CUGUGUCGCAGCUUCUUGUUAGUUGCUUCUUACUAAA-AAGUUGAUUUUUUUCAAACUCACCUUUCAUGGCUGCGACAGUUUGCAAUUGCAA ...((((((((((.(((.(((((........))))).-)))((((......)))).............))))))))))..((((....)))) ( -25.50) >consensus CUGUGUCGCAGCUUCUGGUUACCUGCUUUUUAUUAAA_AAGUUGGUUUUUU_CGAACUCACCUUUCAUGGCUGCGACAGUUUGCAAUUGCAA ...((((((((((...(((............(((((.....))))).............)))......))))))))))..((((....)))) (-18.11 = -18.75 + 0.64)

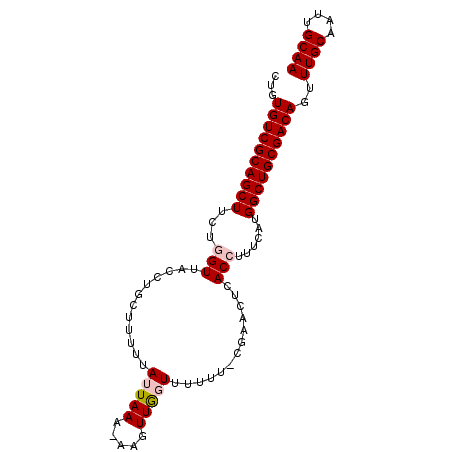

| Location | 18,496,581 – 18,496,672 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 92.32 |
| Mean single sequence MFE | -24.72 |
| Consensus MFE | -17.58 |
| Energy contribution | -17.02 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.16 |
| Mean z-score | -3.47 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.903839 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18496581 91 - 22407834 UUGCAAUUGCAAACUGUCGCAGCCAUGAAAGGUGAGUUCG-AAAAAACUAACUUUUUUAAUAAAAACCAGGUAACCAGAAGCUGCGACACAG ((((....))))..(((((((((...(((((((.((((..-....)))).)))))))............((...))....)))))))))... ( -25.10) >DroSec_CAF1 17081 89 - 1 UUGCAAUUGCAAACUGUCGCAGCCAUGAAAGGUGAGUUCG-AAA-AACCAACUU-UUUAAUAAAAAGCAGGUAACCAGAAGCUGCGACACAG ((((....))))..(((((((((..((((((((..(((..-...-)))..))))-))))..........((...))....)))))))))... ( -25.40) >DroSim_CAF1 16379 90 - 1 UUGCAAUUGCAAACUGUCGCAGCCAUGAAAGGUGAGUUCG-AAAAAACCAAAUU-UUUAAUAAAAAGCAGGUAACCAGAAGCUGCGACACAG ((((....))))..((((((((((((.....)))..(((.-.....(((...((-((.....))))...))).....))))))))))))... ( -21.50) >DroEre_CAF1 17573 90 - 1 UUGCAAUUGCAAACUGUCGCAGCCAUGAAAGGUGAGUUC-AAAAAAAUCAACUU-UUUAGUAAAAAGCAGCCAACAAGAAGCUGCGACACAG ((((....))))..(((((((((..((((((((..(((.-.....)))..))))-))))((.....))............)))))))))... ( -24.20) >DroYak_CAF1 17084 91 - 1 UUGCAAUUGCAAACUGUCGCAGCCAUGAAAGGUGAGUUUGAAAAAAAUCAACUU-UUUAGUAAGAAGCAACUAACAAGAAGCUGCGACACAG ((((....))))..(((((((((..((((((((((.(((....))).)).))))-))))((.((......)).)).....)))))))))... ( -27.40) >consensus UUGCAAUUGCAAACUGUCGCAGCCAUGAAAGGUGAGUUCG_AAAAAACCAACUU_UUUAAUAAAAAGCAGGUAACCAGAAGCUGCGACACAG ((((....))))..(((((((((.......(((.............)))......(((....)))...............)))))))))... (-17.58 = -17.02 + -0.56)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:14:33 2006