| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,493,058 – 18,493,162 |
| Length | 104 |
| Max. P | 0.906735 |

| Location | 18,493,058 – 18,493,162 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 81.29 |
| Mean single sequence MFE | -33.75 |
| Consensus MFE | -22.94 |
| Energy contribution | -23.34 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.906735 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18493058 104 - 22407834 CA-U-UAAUGUGGGUCAACUGCCUGGGCCCGCAGAUGGGAAAGUGUUU------UUCCUAUGUUUUCCGCCGC-UUUUAACGCCUGAUUGCUGCUAAUUGGCGUCAAAAUGGU ((-(-(..((((((((.........))))))))))))((((((..(..------.....)..))))))((((.-((((.(((((.(((((....)))))))))).)))))))) ( -33.80) >DroVir_CAF1 38627 95 - 1 AUCU-UAAUGUAGGUCAAU-GCAUUGAGUU------GGGAAAGUGUU-------U---UAGGUUUUCCGACAGUUUUUGACGCCUAAUGGCUGCUAAUUGGCGUCAAAAUGGC ...(-(((((((......)-)))))))(((------((((((.(...-------.---..).)))))))))...((((((((((.(((........))))))))))))).... ( -31.40) >DroGri_CAF1 15396 103 - 1 AUCUUUAAUGUGGGUCAAC-GCAUUGAGUUGAGUUUGGGAAAGUGUUU------U---UAGGUUUUCCGACACUUUUUGACGCCUGAUUGCUGCUAAUUGGUGUCAAAAUGAC ...(((((((((......)-))))))))..((((((((((((.(....------.---..).)))))))).))))(((((((((.(((((....))))))))))))))..... ( -29.20) >DroSim_CAF1 12730 104 - 1 CA-U-UAAUGUGGGUCAACUGCCUAGGCCCGCAGAUGGGAAAGUGUUU------UUCCUAUAUUUUCCGCCGC-UUUUAACGCCUGAUUGCUGCUAAUUGGCGUCAAAAUGGU ((-(-(..((((((((.........))))))))))))(((((((((..------.....)))))))))((((.-((((.(((((.(((((....)))))))))).)))))))) ( -35.60) >DroEre_CAF1 13995 103 - 1 CA-U-UAAUGUGGGUCAACUGCCUGGGC-CGCAGAUGGGAAAGUGUUU------UUCCUAUGUUUUCCGCCGC-UUUUGACGCCUGAUUGCUGCUAAUUGGCGUCAAAAUGGU ((-(-(..(((((.(((......))).)-))))))))((((((..(..------.....)..))))))((((.-((((((((((.(((((....))))))))))))))))))) ( -37.40) >DroYak_CAF1 13488 107 - 1 CA-U-UAAUGUGGGUCAACUGCCUGGGCCCGCAGAUGGGAAAGUGUUUUUCUUUUUCCUAUGUUUUUCGCCGC-UUUUGACGCCUGAUUGCUGCUAAUUGGCGUCAAAAU--- ..-.-...((((((((.........)))))))).(((((((((.(.....).)))))))))............-((((((((((.(((((....))))))))))))))).--- ( -35.10) >consensus CA_U_UAAUGUGGGUCAACUGCCUGGGCCCGCAGAUGGGAAAGUGUUU______UUCCUAUGUUUUCCGCCGC_UUUUGACGCCUGAUUGCUGCUAAUUGGCGUCAAAAUGGU ........((((((((.........))))))))....((((((..(.............)..))))))......((((((((((.(((((....))))))))))))))).... (-22.94 = -23.34 + 0.39)

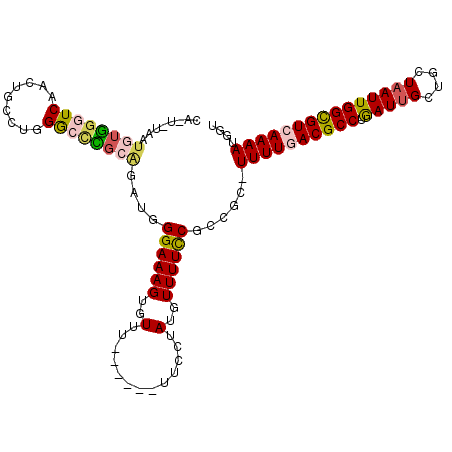

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:14:31 2006