| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,478,449 – 18,478,550 |
| Length | 101 |
| Max. P | 0.741935 |

| Location | 18,478,449 – 18,478,550 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.93 |
| Mean single sequence MFE | -22.60 |
| Consensus MFE | -17.17 |
| Energy contribution | -17.48 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.741935 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
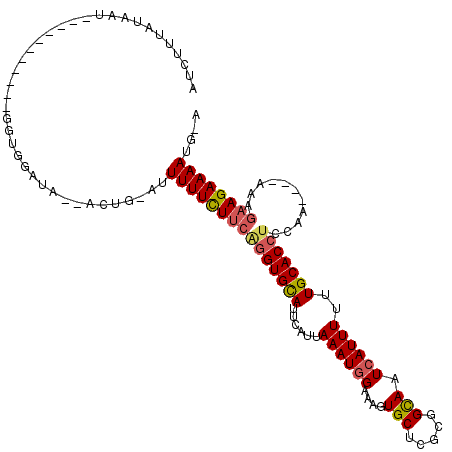
>2L_DroMel_CAF1 18478449 101 + 22407834 AUCUUUAUAAU-----------GGUAGAUA--ACUG-AUUUUUCUUCAGGUGCAUUCAUUAAAUGGAAAGUGCUAGCGGCAAUCAUUUUUUGCACCUCCAA----AAAGAAGAAAAUG-A ((((.......-----------...)))).--....-..(((((((((((((((......((((((....(((.....))).))))))..)))))))....----...))))))))..-. ( -21.31) >DroSec_CAF1 37251 113 + 1 AUCUUUAUAAUGCUUUGGUGUUGGUGGAUA--ACUGGAUUUUUCUUCAGGUGCAUUCAUUAAAUGGAAAGUGCUCGCGGCAAUCAUUUUUUGCACCUCCAA----AAUGAAGAAAAUG-A ((((...((((((....))))))..)))).--.......(((((((((((((((......((((((....(((.....))).))))))..)))))).....----..)))))))))..-. ( -26.81) >DroSim_CAF1 33442 113 + 1 AUCUUUAUAAUGCUUUGGUGUUGGUGGAUA--ACUGGAUUUUUCUUCAGGUGCAUUCAUUAAAUGGAAAGUGCUCGCGGCAAUCAUUUUUUGCACCUCCAA----AAUGAAGAAAAUG-A ((((...((((((....))))))..)))).--.......(((((((((((((((......((((((....(((.....))).))))))..)))))).....----..)))))))))..-. ( -26.81) >DroEre_CAF1 32922 101 + 1 UUGCUUAUAAU-----------GGUGGAUU--ACCA-AUUUUUCUUCGGGUGCAUUCAUUAAAUGGAAAGUGCUCGCGGCAAUCAUUUUUUGCACCUCCAA----AAAGAAGAAAAUG-A ..........(-----------((((...)--))))-..(((((((((((((((......((((((....(((.....))).))))))..)))))))....----...))))))))..-. ( -24.21) >DroYak_CAF1 33291 100 + 1 CAGUUUUUAAU-----------UGUGGACU--ACAG-UUUUUUUUUCAGGUGCAUUCAUUAAAUGGAAAGUGCUCGCGGCAAUCAUUUUUUGCACCUCCAA-----AAGAAGAAAAUG-A .(((((.....-----------...)))))--.((.-(((((((((.(((((((......((((((....(((.....))).))))))..)))))))..))-----)))))))...))-. ( -22.50) >DroAna_CAF1 26241 91 + 1 --------------------------GACUCUAUCC-CUUUUUCUC--UGUGUAAUCAUUAAAUGGAAAGUGCUGGCUGUAAUAAUUUUUUGCACCUCCAAAGAAAAAGAAGAAAAUGUA --------------------------..........-((((((((.--.(((....)))....((((..((((..................)))).)))).))))))))........... ( -13.97) >consensus AUCUUUAUAAU___________GGUGGAUA__ACUG_AUUUUUCUUCAGGUGCAUUCAUUAAAUGGAAAGUGCUCGCGGCAAUCAUUUUUUGCACCUCCAA____AAAGAAGAAAAUG_A .......................................(((((((((((((((......((((((....(((.....))).))))))..)))))))...........)))))))).... (-17.17 = -17.48 + 0.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:14:27 2006