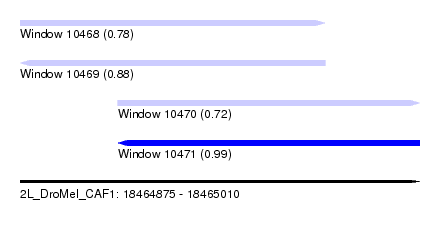
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,464,875 – 18,465,010 |
| Length | 135 |
| Max. P | 0.987309 |

| Location | 18,464,875 – 18,464,978 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 91.65 |
| Mean single sequence MFE | -31.06 |
| Consensus MFE | -24.44 |
| Energy contribution | -24.92 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.781968 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18464875 103 + 22407834 GCAGAAAGAGCUCAGUGUAUUGCAUAAUUUCGUGCAACUCCCACAAACAUAGGCGUUUUUGAUCUGUCUGUGUGUAGGUUCCUCGCUCUGCAUGUGUAUGCGU (((((..(((....(((..((((((......))))))....)))..(((((((((.........)))))))))........)))..)))))((((....)))) ( -27.50) >DroSec_CAF1 19311 103 + 1 GCACAAAGAGUUCAGCGUAUUGCAUAAUUUCGUGCAACUCCAACACACAUAGACGCUUUUGAUCUGUCUGUGUGUAGGUUCCUCGCUCUGCAUGUGUAUGCGA (((((.(((((..(((...((((((......)))))).......(((((((((((.........)))))))))))..)))....)))))...)))))...... ( -29.50) >DroSim_CAF1 19720 103 + 1 GCACAAAGAGUUCAGCGUAUUGCAUAAUUUCGUGCAACUCCAACACACAUAGGCGCUUUUGAUCUGUCUGUGUGUAGGUUCCUCGCUCUGCAUGUGUAUGCGU (((((.(((((..(((...((((((......)))))).......(((((((((((.........)))))))))))..)))....)))))...)))))...... ( -28.90) >DroEre_CAF1 19159 103 + 1 GCACAAAGAGCUUAGCGUAUUGCAUAAUUUCGUGCAACUCCCACACAGAUAGGCGCUUCUGAUCUGUCUGCGUGUAGGUUCCUCGCUCUGCAUGUGUAUGCGU (((((.(((((..........((((......))))((((..(((.((((((((((....)).)))))))).)))..))))....)))))...)))))...... ( -33.20) >DroYak_CAF1 19423 103 + 1 GUACUAAGAGCUUAGCGUAUUGCAUAAUUUUGUGCAACUCCCACACACAUAGGCGCUUUUGGCCUGUCUGUGUGGAGGUUCCUCGUUCUGCAUGUGUAUGCAU ......(((((..........(((((....)))))(((..(((((((.((((((.(....))))))).)))))))..)))....)))))((((....)))).. ( -36.20) >consensus GCACAAAGAGCUCAGCGUAUUGCAUAAUUUCGUGCAACUCCCACACACAUAGGCGCUUUUGAUCUGUCUGUGUGUAGGUUCCUCGCUCUGCAUGUGUAUGCGU (((((.(((((........((((((......))))))..((...(((((((((((.........))))))))))).))......)))))...)))))...... (-24.44 = -24.92 + 0.48)

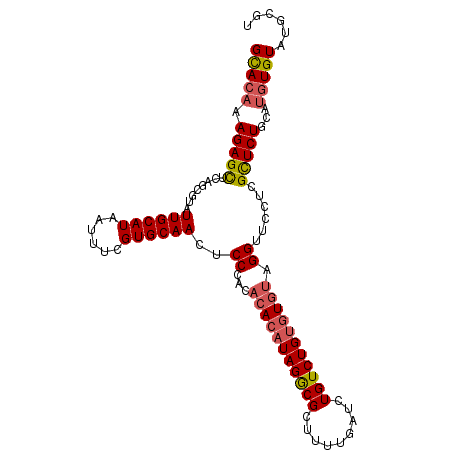
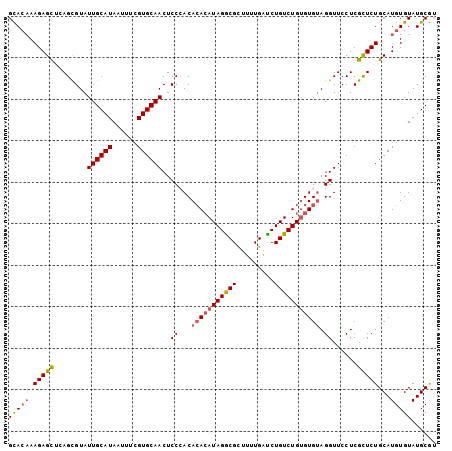
| Location | 18,464,875 – 18,464,978 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 91.65 |
| Mean single sequence MFE | -28.32 |
| Consensus MFE | -23.84 |
| Energy contribution | -24.92 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.883604 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18464875 103 - 22407834 ACGCAUACACAUGCAGAGCGAGGAACCUACACACAGACAGAUCAAAAACGCCUAUGUUUGUGGGAGUUGCACGAAAUUAUGCAAUACACUGAGCUCUUUCUGC ..((((....))))(((((..((..((((((.(((....(........).....))).)))))).((((((........))))))...))..)))))...... ( -26.20) >DroSec_CAF1 19311 103 - 1 UCGCAUACACAUGCAGAGCGAGGAACCUACACACAGACAGAUCAAAAGCGUCUAUGUGUGUUGGAGUUGCACGAAAUUAUGCAAUACGCUGAACUCUUUGUGC ..((((....))))...(((((((.((.(((((((...((((.......)))).))))))).)).((((((........)))))).........))))))).. ( -28.60) >DroSim_CAF1 19720 103 - 1 ACGCAUACACAUGCAGAGCGAGGAACCUACACACAGACAGAUCAAAAGCGCCUAUGUGUGUUGGAGUUGCACGAAAUUAUGCAAUACGCUGAACUCUUUGUGC ..((((....))))...(((((((.((.(((((((....(........).....))))))).)).((((((........)))))).........))))))).. ( -25.80) >DroEre_CAF1 19159 103 - 1 ACGCAUACACAUGCAGAGCGAGGAACCUACACGCAGACAGAUCAGAAGCGCCUAUCUGUGUGGGAGUUGCACGAAAUUAUGCAAUACGCUAAGCUCUUUGUGC ..(((((.....((..((((.....((..((((((((.((...........)).)))))))))).((((((........)))))).))))..))....))))) ( -29.70) >DroYak_CAF1 19423 103 - 1 AUGCAUACACAUGCAGAACGAGGAACCUCCACACAGACAGGCCAAAAGCGCCUAUGUGUGUGGGAGUUGCACAAAAUUAUGCAAUACGCUAAGCUCUUAGUAC .(((((....)))))......(.(((..(((((((.(.(((((....).)))).).)))))))..))).).................(((((....))))).. ( -31.30) >consensus ACGCAUACACAUGCAGAGCGAGGAACCUACACACAGACAGAUCAAAAGCGCCUAUGUGUGUGGGAGUUGCACGAAAUUAUGCAAUACGCUGAGCUCUUUGUGC ..((((....))))...(((((((.((((((((((...((...........)).)))))))))).((((((........)))))).........))))))).. (-23.84 = -24.92 + 1.08)

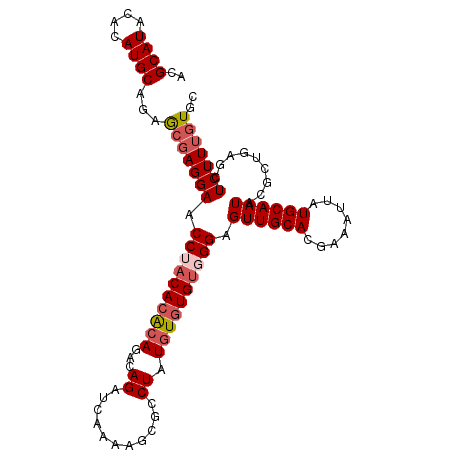
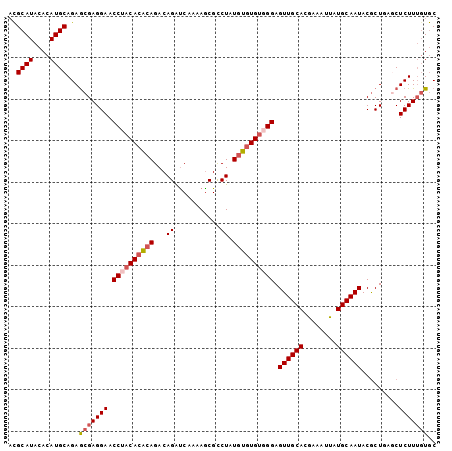
| Location | 18,464,908 – 18,465,010 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 92.84 |
| Mean single sequence MFE | -35.02 |
| Consensus MFE | -27.20 |
| Energy contribution | -26.88 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.722016 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18464908 102 + 22407834 GCAACUCCCACAAACAUAGGCGUUUUUGAUCUGUCUGUGUGUAGGUUCCUCGCUCUGCAUGUGUAUGCGUGUGUGGCAUUGGCUUUGAUUGUGUGUAGGAGC ....((((((((((((((((((.........)))))))))((..((.((.(((.(.((((....))))).))).)).))..)).....)))))....)))). ( -27.90) >DroSec_CAF1 19344 102 + 1 GCAACUCCAACACACAUAGACGCUUUUGAUCUGUCUGUGUGUAGGUUCCUCGCUCUGCAUGUGUAUGCGAGUGUGGCAUUGGCUUUGAUUGUGUGUAGGAGC ....((((.(((((((..((.(((..(((((((.(.....)))))).((.(((((.((((....))))))))).))))..))).))...))))))).)))). ( -35.70) >DroSim_CAF1 19753 102 + 1 GCAACUCCAACACACAUAGGCGCUUUUGAUCUGUCUGUGUGUAGGUUCCUCGCUCUGCAUGUGUAUGCGUGUGUGACAUUGGCUUUGAUUGUGUGUAGGAGC ....((((.(((((((..((.(((..((.((.....(.(((.((....))))).).((((((....))))))..))))..))).))...))))))).)))). ( -31.00) >DroEre_CAF1 19192 101 + 1 GCAACUCCCACACAGAUAGGCGCUUCUGAUCUGUCUGCGUGUAGGUUCCUCGCUCUGCAUGUGUAUGCGUGUGUGGCAUUGGCU-UGUUUGUGUGUAGGAGC ....(((((((((((((((((......((((((.(.....)))))))....(((..((((((....))))))..)))....)))-))))))))))..)))). ( -36.90) >DroYak_CAF1 19456 102 + 1 GCAACUCCCACACACAUAGGCGCUUUUGGCCUGUCUGUGUGGAGGUUCCUCGUUCUGCAUGUGUAUGCAUGUGUGGCAUUGGCUUUGCUUGUGUGUGGGAGC ....((((((((((((.((((.(....)))))(((.((((.(((....))).....(((((((....))))))).)))).)))......)))))))))))). ( -43.60) >consensus GCAACUCCCACACACAUAGGCGCUUUUGAUCUGUCUGUGUGUAGGUUCCUCGCUCUGCAUGUGUAUGCGUGUGUGGCAUUGGCUUUGAUUGUGUGUAGGAGC ....((((.(((((((..((.(((..((.((.....(.(((.((....))))).).(((..((....))..)))))))..))).))...))))))).)))). (-27.20 = -26.88 + -0.32)

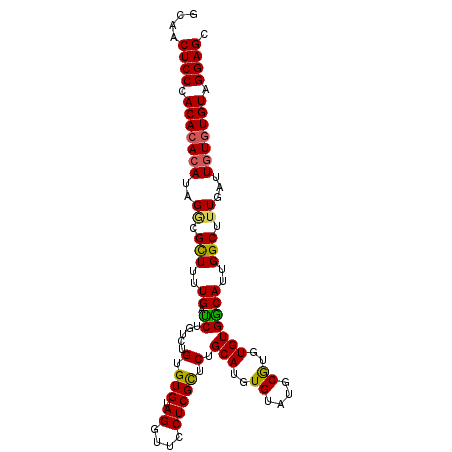
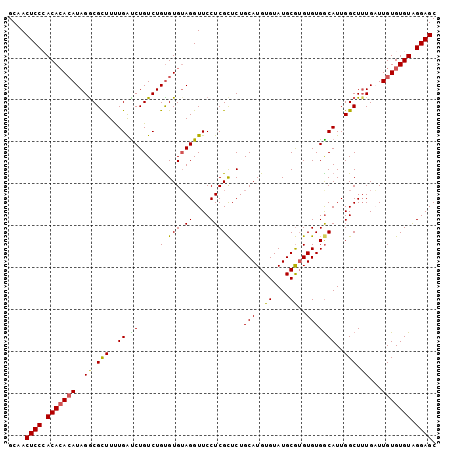
| Location | 18,464,908 – 18,465,010 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 92.84 |
| Mean single sequence MFE | -27.99 |
| Consensus MFE | -22.98 |
| Energy contribution | -23.78 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.987309 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18464908 102 - 22407834 GCUCCUACACACAAUCAAAGCCAAUGCCACACACGCAUACACAUGCAGAGCGAGGAACCUACACACAGACAGAUCAAAAACGCCUAUGUUUGUGGGAGUUGC (((((((((.(((.....................((((....))))((.(((((....)).......((....)).....))))).))).)))))))))... ( -22.90) >DroSec_CAF1 19344 102 - 1 GCUCCUACACACAAUCAAAGCCAAUGCCACACUCGCAUACACAUGCAGAGCGAGGAACCUACACACAGACAGAUCAAAAGCGUCUAUGUGUGUUGGAGUUGC (((((.(((((((.............((.(.(((((((....)))).))).).))...........((((...........)))).))))))).)))))... ( -28.80) >DroSim_CAF1 19753 102 - 1 GCUCCUACACACAAUCAAAGCCAAUGUCACACACGCAUACACAUGCAGAGCGAGGAACCUACACACAGACAGAUCAAAAGCGCCUAUGUGUGUUGGAGUUGC (((((.(((((((......((...((((......((((....))))...(.(((....)).).)...))))........)).....))))))).)))))... ( -25.30) >DroEre_CAF1 19192 101 - 1 GCUCCUACACACAAACA-AGCCAAUGCCACACACGCAUACACAUGCAGAGCGAGGAACCUACACGCAGACAGAUCAGAAGCGCCUAUCUGUGUGGGAGUUGC (((((((((((......-.((...((........((((....))))...(((.(.......).)))........))...)).......)))))))))))... ( -26.04) >DroYak_CAF1 19456 102 - 1 GCUCCCACACACAAGCAAAGCCAAUGCCACACAUGCAUACACAUGCAGAACGAGGAACCUCCACACAGACAGGCCAAAAGCGCCUAUGUGUGUGGGAGUUGC (((((((((((((((....((....(((.....(((((....)))))....(((....)))..........))).....))..)).)))))))))))))... ( -36.90) >consensus GCUCCUACACACAAUCAAAGCCAAUGCCACACACGCAUACACAUGCAGAGCGAGGAACCUACACACAGACAGAUCAAAAGCGCCUAUGUGUGUGGGAGUUGC (((((((((((((.....................((((....))))((.(((((....)).......(......).....))))).)))))))))))))... (-22.98 = -23.78 + 0.80)

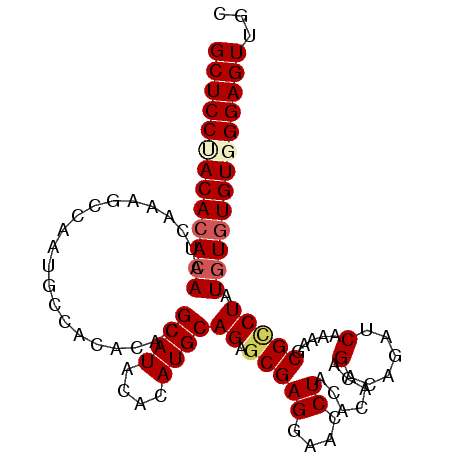
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:14:16 2006