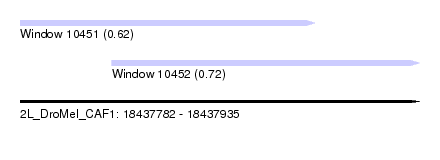
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,437,782 – 18,437,935 |
| Length | 153 |
| Max. P | 0.717107 |

| Location | 18,437,782 – 18,437,895 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 86.35 |
| Mean single sequence MFE | -17.53 |
| Consensus MFE | -12.75 |
| Energy contribution | -13.53 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.617625 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
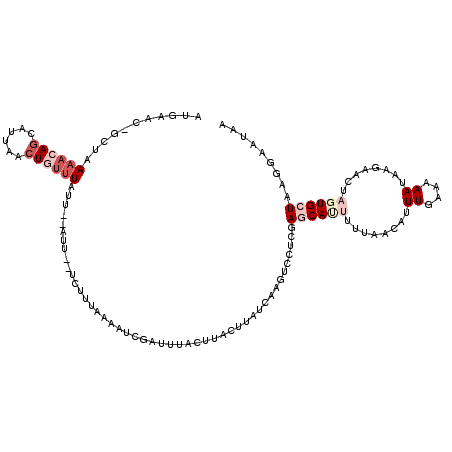
>2L_DroMel_CAF1 18437782 113 + 22407834 CACCCAAUAGCAGUUUUCUCCAUUAACUUUGGUAUAUGAACAACUAAAACAGUAUUAACUGUUUAUUUUAUUACUGUACUUACUAUUUACUUACUUAUCAAGUCCUCGAGCAU .........((.((((...(((.......))).....)))).....(((((((....))))))).............................................)).. ( -12.70) >DroSec_CAF1 276335 111 + 1 CACCCAAUAACAGUUUUCUCCAUUAACUUUGGUAUAUGAACUGCUAAAACAGCAUUAACUGUUUAUU--AUUUCUUUAUAAUGGGUUUACUUACCUAUUAAGUCCUAGAUCAC ..........((((((...(((.......))).....))))))(((((((((......))))))...--.........((((((((......)))))))).....)))..... ( -20.30) >DroSim_CAF1 290141 111 + 1 CACCCUAUAGCAGUUUUCUCUAUUAACUUUGGUAUAUGAACUGCUAAAACAGCAUUAACUGUUUAUU--AUUUCUUUAUAAUGGAUUUACUUACCUAUCAAGUACUAGAUCAC .......(((((((((...(((.......))).....)))))))))((((((......))))))(((--((......))))).((((((..(((.......))).)))))).. ( -19.60) >consensus CACCCAAUAGCAGUUUUCUCCAUUAACUUUGGUAUAUGAACUGCUAAAACAGCAUUAACUGUUUAUU__AUUUCUUUAUAAUGGAUUUACUUACCUAUCAAGUCCUAGAUCAC .......(((((((((...(((.......))).....)))))))))((((((......))))))................................................. (-12.75 = -13.53 + 0.78)

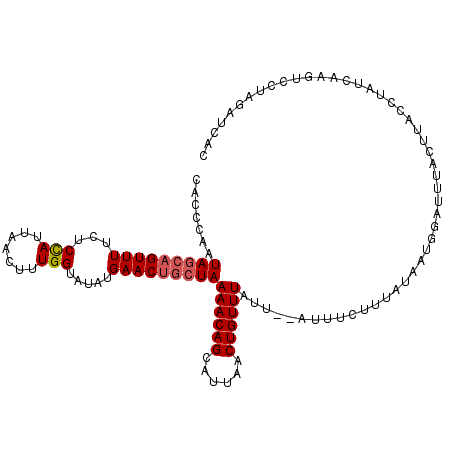
| Location | 18,437,817 – 18,437,935 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.67 |
| Mean single sequence MFE | -18.81 |
| Consensus MFE | -0.89 |
| Energy contribution | -3.91 |
| Covariance contribution | 3.02 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.05 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.717107 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18437817 118 + 22407834 AUGAACAACUAAAACAGUAUUAACUGUUUAUUUUAUU--ACUGUACUUACUAUUUACUUACUUAUCAAGUCCUCGAGCAUUUUUAACAUUUGAAAAAUAAGAACCAGUGCUAAGGAAUAA ...........(((((((....)))))))........--..............................((((..((((((........((....))........)))))).)))).... ( -15.09) >DroSec_CAF1 276370 116 + 1 AUGAACUGCUAAAACAGCAUUAACUGUUUAUU--AUU--UCUUUAUAAUGGGUUUACUUACCUAUUAAGUCCUAGAUCACUUUUAGCAUUUGCUAAAUAAGAACUAGUGCUAAGGAAUAU ...........((((((......))))))..(--(((--((((..((((((((......))))))))((..((((.((...((((((....))))))...)).))))..)))))))))). ( -31.00) >DroSim_CAF1 290176 116 + 1 AUGAACUGCUAAAACAGCAUUAACUGUUUAUU--AUU--UCUUUAUAAUGGAUUUACUUACCUAUCAAGUACUAGAUCACUUUUAGCAUUUGAUAAAUAAGAGCUAGUGCUAAGGAAUAA ...............(((((((.(((((((((--((.--.....)))))))))...((((..((((((((.(((((.....))))).))))))))..)))))).)))))))......... ( -25.20) >DroEre_CAF1 276709 96 + 1 AU-----GCAAAAUCAGCAUUAACUCCCUAUU--AUUUUACCUUAAACUCAC----CUUACUUAUCCAGUCCUCGAGCAUGUUUAACAAUUGGAAAAUAA------UUGCUAA------- ..-----........((((..........(((--(((((.((((((((....----((((((.....)))....)))...)))))).....)))))))))------)))))..------- ( -8.50) >DroYak_CAF1 291702 105 + 1 AU-----UCAAAAACAGCAUUAAGUUUUUAUU--AUUAUUCCUUAAACUCGC----CUCACUUAUCCA----UCGAGCAUUUCUAACAUUUGGAAAAAAAGAACUUUUGCUAAGGAAUCA ..-----........((((..((((((((...--.............((((.----............----.))))..((((((.....))))))..)))))))).))))......... ( -14.24) >consensus AUGAAC_GCUAAAACAGCAUUAACUGUUUAUU__AUU__UCUUUAAAAUCGAUUUACUUACUUAUCAAGUCCUCGAGCAUUUUUAACAUUUGAAAAAUAAGAACUAGUGCUAAGGAAUAA ...........((((((......))))))..............................................((((((........((....))........))))))......... ( -0.89 = -3.91 + 3.02)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:13:58 2006