
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,433,854 – 18,434,004 |
| Length | 150 |
| Max. P | 0.967600 |

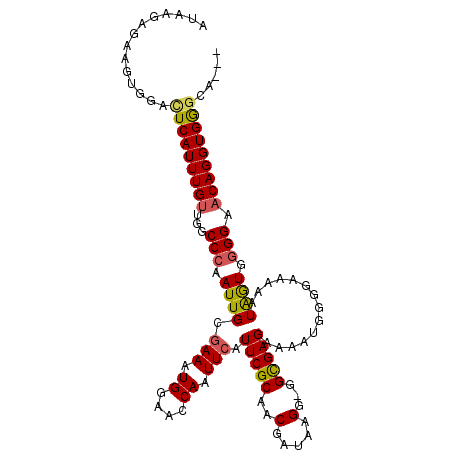
| Location | 18,433,854 – 18,433,964 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 90.15 |
| Mean single sequence MFE | -27.40 |
| Consensus MFE | -20.04 |
| Energy contribution | -20.00 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.797135 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18433854 110 - 22407834 AUAAGAGAAGUGGACUCAUUUGUUGGCCCAAUUGCGAAAUGGAACCAAUUCAUUCGCAACGAUAAGG-GGCGAGAAAAUGGGGAAAAAUAGUGGGGAACAGGUGGGCAGGU ............(.((((((((((..((((.....(((.((....)).))).(((((..(.....).-.))))).................)))).))))))))))).... ( -26.90) >DroSec_CAF1 272290 107 - 1 AUAAGAGUAGUGGGUUCAUUUGUUGGCCCAAUUGCGAAAUGGAACCAAUUCAUUCGCAACGAUAAGG-GGCGAGAAAAUGGGGAAAAAUAGUGGGGAACAGGUGGGCA--- .............(((((((((((..((((.....(((.((....)).))).(((((..(.....).-.))))).................)))).))))))))))).--- ( -28.20) >DroSim_CAF1 286162 107 - 1 AUAAGAGAAGUGGACUCAUUUGUUGGCCCAAUUGCGAAAUGGAACCAAUUCAUUCGCAACGAUAAGG-GGCGAGAAAAUGGGGAAAAAUAGUGGGGAACAGGUGGGCA--- ............(.((((((((((..((((.....(((.((....)).))).(((((..(.....).-.))))).................)))).))))))))))).--- ( -26.90) >DroEre_CAF1 272929 107 - 1 AUACAAGAAAUGGACUCAUUUGCUGGCCCAAUUGCGAAAUGGAACCAAUUCAUUCGCAUCGAUAAGG-GGCGAGAAAGUGGGGAAAACUGAUGUGGAACAGGUGAACA--- .(((..........(((((((.((.((((...((((((.((((.....))))))))))........)-))).)).))))))).....(((........))))))....--- ( -28.90) >DroYak_CAF1 287488 108 - 1 AUACAAAAAUUGGACUCAUUUGUUGGCCCAAUUGCAAAAUGGAACCAAUUCAUUCGCAUCGAAAGGGUGGUGAGAAAAUGGGGAAAAAUAGUGGGGAACAGGUGAGCA--- ............(.((((((((((..((((.((.(.....).))((.(((..(((((.((....))...)))))..))).)).........)))).))))))))))).--- ( -26.10) >consensus AUAAGAGAAGUGGACUCAUUUGUUGGCCCAAUUGCGAAAUGGAACCAAUUCAUUCGCAACGAUAAGG_GGCGAGAAAAUGGGGAAAAAUAGUGGGGAACAGGUGGGCA___ ..............(((((((((...(((.((((.(((.((....)).))).(((((..(.....)...)))))..............)))).))).)))))))))..... (-20.04 = -20.00 + -0.04)


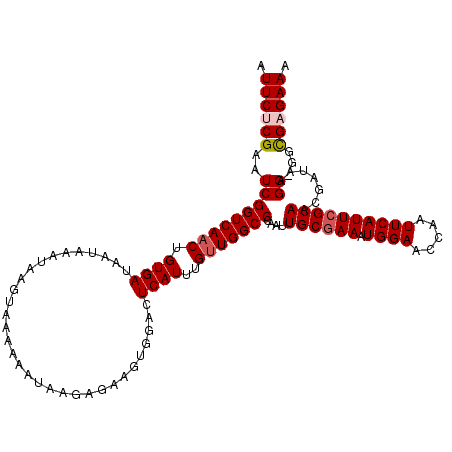
| Location | 18,433,888 – 18,434,004 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 93.04 |
| Mean single sequence MFE | -27.83 |
| Consensus MFE | -22.59 |
| Energy contribution | -23.23 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967600 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18433888 116 - 22407834 AUUCUCGAAUCGGUCAACUGUGAUAAUAAAUAAGUAAAAAAUAAGAGAAGUGGACUCAUUUGUUGGCCCAAUUGCGAAAUGGAACCAAUUCAUUCGCAACGAUAAGG-GGCGAGAAA .((((((..((((((((((((....)))................(((.......)))....)))))))...(((((((.((((.....)))))))))))......))-..)))))). ( -29.80) >DroSec_CAF1 272321 116 - 1 AUUCUCGAAUCGGUCAACUGUGAUAAUAAAUACGUAAAAAAUAAGAGUAGUGGGUUCAUUUGUUGGCCCAAUUGCGAAAUGGAACCAAUUCAUUCGCAACGAUAAGG-GGCGAGAAA .((((((..(((((((((.((((.......(((.............)))......))))..)))))))...(((((((.((((.....)))))))))))......))-..)))))). ( -32.94) >DroSim_CAF1 286193 116 - 1 AUUCUCGAAUCGGUCAACUGUGAUAAUAAAUACGUAAAAAAUAAGAGAAGUGGACUCAUUUGUUGGCCCAAUUGCGAAAUGGAACCAAUUCAUUCGCAACGAUAAGG-GGCGAGAAA .((((((..(((((((((.((((.......(((................)))...))))..)))))))...(((((((.((((.....)))))))))))......))-..)))))). ( -30.49) >DroEre_CAF1 272960 115 - 1 AUUCCCGAAUCGGUCAACUGUGAUAAUAAAUAAGUAAA-AAUACAAGAAAUGGACUCAUUUGCUGGCCCAAUUGCGAAAUGGAACCAAUUCAUUCGCAUCGAUAAGG-GGCGAGAAA .....(((((.((((...(((....))).....(((..-..)))........)))).)))))((.((((...((((((.((((.....))))))))))........)-))).))... ( -24.70) >DroYak_CAF1 287519 117 - 1 AUUCCCGAAUCGGUCAACUGUGAUAAUAAAUAAGUAAAAAAUACAAAAAUUGGACUCAUUUGUUGGCCCAAUUGCAAAAUGGAACCAAUUCAUUCGCAUCGAAAGGGUGGUGAGAAA ...(((...(((((((((.((((...(((....(((.....))).....)))...))))..)))))))....(((..((((((.....)))))).)))..))..))).......... ( -21.20) >consensus AUUCUCGAAUCGGUCAACUGUGAUAAUAAAUAAGUAAAAAAUAAGAGAAGUGGACUCAUUUGUUGGCCCAAUUGCGAAAUGGAACCAAUUCAUUCGCAACGAUAAGG_GGCGAGAAA .((((((..(((((((((.((((................................))))..)))))))....((((((.((((.....)))))))))).......))...)))))). (-22.59 = -23.23 + 0.64)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:13:55 2006