| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,419,024 – 18,419,129 |
| Length | 105 |
| Max. P | 0.843645 |

| Location | 18,419,024 – 18,419,129 |
|---|---|
| Length | 105 |
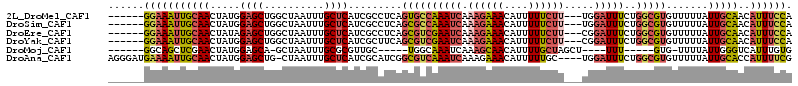
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 81.54 |
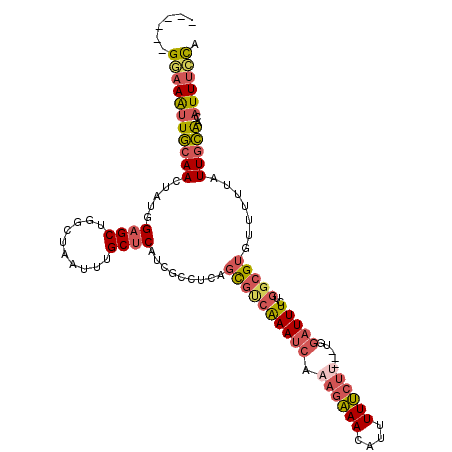
| Mean single sequence MFE | -29.25 |
| Consensus MFE | -18.45 |
| Energy contribution | -19.15 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.843645 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18419024 105 - 22407834 ------GGAAAUUGCAACUAUGGAGCUGGCUAAUUUGCUCAUCGCCUCAGUGCCAAAUCAAAGAAACAUUUUUCUU---UGGAUUUCUGGCGUGUUUUUAUUGCAACAUUUCCA ------(((((((((((((..((.(.((((......)).)).).))..)))(((((((((((((((....))))))---).))))..))))..........))))..)))))). ( -29.60) >DroSim_CAF1 271611 105 - 1 ------GGAAAUUGCAACUAUGGAGCUGGCUAAUUUGCUCAUCGCCUCAGCGCCAAAUCAAAGAAACAUUUUUCUU---UGGAUUUCUGGCGUGUUUUUAUUGCAACAUUUCCA ------(((((((((((...((..((((((......)).))..))..))(((((((((((((((((....))))))---).))))..)))))).......)))))..)))))). ( -32.40) >DroEre_CAF1 258247 105 - 1 ------GGAAAUUGCAACUAUAGAGCUGGCUAAUUUGCUCAUCGCCUCAGCGUCGAAUCAAAGAAACAUUUUUCUU---CGGAUUUCUGGCGUGUUUUUAUUGCAACAUUUCCA ------(((((((((((.....((((..........)))).........((((((((((.((((((....))))))---..)))))..))))).......)))))..)))))). ( -27.20) >DroYak_CAF1 271711 105 - 1 ------GGAAAUUGCAACUAUGGAGCUGGCUAAUUUGCUCAUCGCUUCAGCGUCGAAUCAAAGAAACAUUUUUCUU---CGGAUUUCUGGCGUGUUUUUAUUGCAACAUUUCCA ------(((((((((((...((((((((((......)).))..))))))((((((((((.((((((....))))))---..)))))..))))).......)))))..)))))). ( -31.20) >DroMoj_CAF1 405136 92 - 1 ------GGCAGCUCGAACUAUGGAGCA-GCUAAUUUGCGCGUUGC-----UGGCAAAUCAAAGCAACAUUUUGCUAGCU----UUU-----GUG-UUUUAUUGGGUCAUUUGUG ------(((.((((........)))).-.(((((..(((((..((-----(((((((............))))))))).----..)-----)))-)...))))))))....... ( -29.40) >DroAna_CAF1 237626 109 - 1 AGGGAUGAAAAUUGCAACUAUGGAGCUG-CUAAUUUGCUCAUCGCAUCGGCGUCAAAUCAAAGAAACAUUUUUGC----UGGAUUUCUGGCGUGUUUUUAUUGCACCAUUUUCG ......(((((((((((...((..((((-......(((.....))).))))..))......((((((((....((----..(....)..)))))))))).)))))..)))))). ( -25.70) >consensus ______GGAAAUUGCAACUAUGGAGCUGGCUAAUUUGCUCAUCGCCUCAGCGUCAAAUCAAAGAAACAUUUUUCUU___UGGAUUUCUGGCGUGUUUUUAUUGCAACAUUUCCA ......(((((((((((.....((((..........)))).........((((((((((.((((((....)))))).....)))))..))))).......)))))..)))))). (-18.45 = -19.15 + 0.70)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:13:46 2006