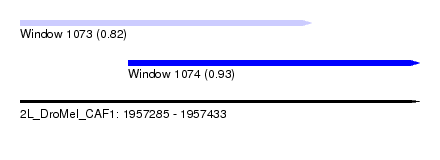
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,957,285 – 1,957,433 |
| Length | 148 |
| Max. P | 0.929336 |

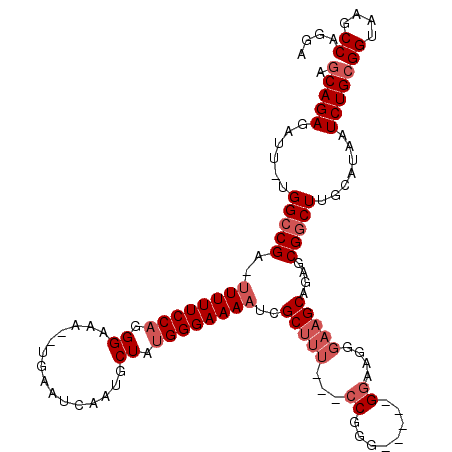
| Location | 1,957,285 – 1,957,393 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.59 |
| Mean single sequence MFE | -36.58 |
| Consensus MFE | -14.30 |
| Energy contribution | -16.78 |
| Covariance contribution | 2.48 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.39 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.819786 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1957285 108 + 22407834 ACCUCGGGAAGGAACUGAACUUGGGCUGAUGCCCGAUUGCAGCAGAGAUU-UGGCCGA-UUUUUCCAGGGAAA--UGAAUCAAUGCUAUGGGAAAAUCGCUUU---CCGGG-----GGAA .((((((((((...(((...((((((....))))))...)))........-....(((-((((((((.((...--((...))...)).)))))))))))))))---)))))-----)... ( -42.40) >DroSim_CAF1 1672 108 + 1 ACCUCGGGAAGGAACUGAACUUGGGCUGAUGCCAGAUUGCAGCAGAGAUU-UGGCCGA-UUUUUCCAGGGAAA--UGAAUCAAUGCUAUGGGAAAAUCGCUUU---CCGGG-----GGAA .((((((((((...................((((((((........))))-))))(((-((((((((.((...--((...))...)).)))))))))))))))---)))))-----)... ( -38.90) >DroEre_CAF1 8979 109 + 1 ACCUCGGCCAGGAACUGAGCUUGGGCUGAUGCCAGAUUGCAGCAGAGAUUUUGGACGA-UUUUUCCAGGGAAA--UGAAUCAAUGCUAUGGGAAAAUCGCUUU---CCGGG-----GGAA .((((((((((((.((..(((..(((....))).......)))..)).)))))).(((-((((((((.((...--((...))...)).)))))))))))....---)))))-----)... ( -36.00) >DroYak_CAF1 6662 116 + 1 ACCUCGCUGAGGAACUGAGCUUGGGCUGAUGCCAGAUUGCAGCAGAGAUU-UGGCCGA-UUUUUCCAGGGAAA--UGAAUCAAUGCUAUGGGAAAAUCGCUUUUCCCCGGUGAAAGGAAG .((((((((.((((..((((..........((((((((........))))-)))).((-((((((((.((...--((...))...)).)))))))))))))))))).))))))..))... ( -38.30) >DroAna_CAF1 29375 92 + 1 ACCUCCGGGA----CGGAGAGAGUCCUGAUGCCAGAUUGCAACAGAGAUU-UGGCCGAGUUUUUCCUUGGAAAAUCGAAUCAAUUCUAUGGGAAAU-----------------------A ..((((((((----(.......))))))..((((((((........))))-)))).))).((((((.(((((...........))))).)))))).-----------------------. ( -27.30) >consensus ACCUCGGGAAGGAACUGAGCUUGGGCUGAUGCCAGAUUGCAGCAGAGAUU_UGGCCGA_UUUUUCCAGGGAAA__UGAAUCAAUGCUAUGGGAAAAUCGCUUU___CCGGG_____GGAA ..(((((.......)))))(((((.(((.(((......))).)))..........(((.((((((((.((...............)).))))))))))).......)))))......... (-14.30 = -16.78 + 2.48)

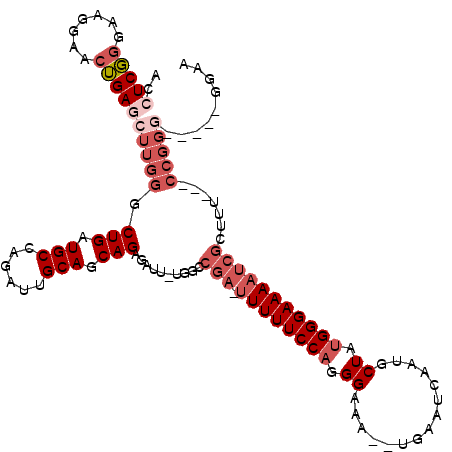
| Location | 1,957,325 – 1,957,433 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.78 |
| Mean single sequence MFE | -35.46 |
| Consensus MFE | -21.30 |
| Energy contribution | -23.10 |
| Covariance contribution | 1.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929336 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1957325 108 + 22407834 AGCAGAGAUU-UGGCCGA-UUUUUCCAGGGAAA--UGAAUCAAUGCUAUGGGAAAAUCGCUUU---CCGGG-----GGAAGGGAAGCAGAGCGGCUUGCAUAAUCUGCGGUAAGCCAGGA .(((((....-.((((((-((((((((.((...--((...))...)).))))))))))(((((---((...-----....))))))).....)))).......)))))((....)).... ( -39.10) >DroSim_CAF1 1712 108 + 1 AGCAGAGAUU-UGGCCGA-UUUUUCCAGGGAAA--UGAAUCAAUGCUAUGGGAAAAUCGCUUU---CCGGG-----GGAAGGGAAGCAGAGCGGCUUGCAUAAUCUGCGGUAAGCCAGGA .(((((....-.((((((-((((((((.((...--((...))...)).))))))))))(((((---((...-----....))))))).....)))).......)))))((....)).... ( -39.10) >DroEre_CAF1 9019 109 + 1 AGCAGAGAUUUUGGACGA-UUUUUCCAGGGAAA--UGAAUCAAUGCUAUGGGAAAAUCGCUUU---CCGGG-----GGAAGCGAAGCAGAGCGGCUUGCAUAAUCUGCGGUAAGCCAGGA .(((((..((((((((((-((((((((.((...--((...))...)).)))))))))))...)---)))))-----)...((.((((......))))))....)))))((....)).... ( -33.30) >DroYak_CAF1 6702 116 + 1 AGCAGAGAUU-UGGCCGA-UUUUUCCAGGGAAA--UGAAUCAAUGCUAUGGGAAAAUCGCUUUUCCCCGGUGAAAGGAAGGGGAAGCAGAGCGGCUUGCAUAAUCUGCGGUAAGCCAGAA .(((((....-.((((((-((((((((.((...--((...))...)).))))))))))((((((((((...........))))))...)))))))).......)))))((....)).... ( -41.70) >DroAna_CAF1 29411 92 + 1 AACAGAGAUU-UGGCCGAGUUUUUCCUUGGAAAAUCGAAUCAAUUCUAUGGGAAAU-----------------------AGAGAA----CACGGCUUGCAUAAUCUGCGGUAAGCCAGAA ......((((-((((((((......)))))....)))))))..((((((.....))-----------------------))))..----...(((((((..........))))))).... ( -24.10) >consensus AGCAGAGAUU_UGGCCGA_UUUUUCCAGGGAAA__UGAAUCAAUGCUAUGGGAAAAUCGCUUU___CCGGG_____GGAAGGGAAGCAGAGCGGCUUGCAUAAUCUGCGGUAAGCCAGGA .(((((......(((((..((((((((.((...............)).))))))))..(((((...((........))....)))))....))))).......)))))((....)).... (-21.30 = -23.10 + 1.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:42:07 2006