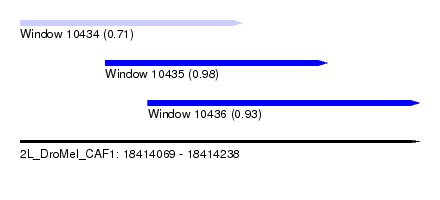
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,414,069 – 18,414,238 |
| Length | 169 |
| Max. P | 0.976880 |

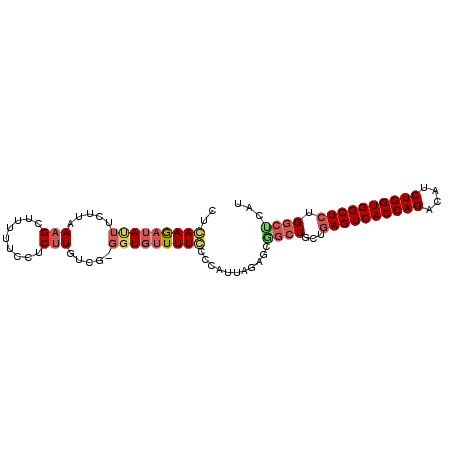
| Location | 18,414,069 – 18,414,163 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 90.07 |
| Mean single sequence MFE | -31.21 |
| Consensus MFE | -20.38 |
| Energy contribution | -21.54 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.30 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.709510 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18414069 94 + 22407834 GUCAAGAUAUUUCUUAAAGCUUUUUCCUCUUGUCG-GUUGUUUUCCCCAUUAGAGCAGCUGCUGAGUCACCAGACAUCUGGUGGCUCUGGCUCAU (.(((((....................))))).)(-((((((((.......)))))))))(((((((((((((....)))))))))).))).... ( -30.95) >DroSec_CAF1 254673 94 + 1 CUCAAGAUACUUCUUAAAGCUUUUUCCUCUUCUCG-GGUGUUUUGCCCAUUAGAGCGGCUGCUGAGUCACCAGACAUCUGGUGGCUCUGGCUCAU ...((((....))))..((((.....((((....(-(((.....))))...)))).))))(((((((((((((....)))))))))).))).... ( -34.00) >DroSim_CAF1 266329 94 + 1 CUCAAGAAAUUUCUUAAAGCUUUUUCCUCUUCUCG-GGUGUUUUGCCCAUUAGAGCGGCUGCUGAGUCACCAGACAUCUGGUGGCUCUGGCUCAU ...((((....))))..((((.....((((....(-(((.....))))...)))).))))(((((((((((((....)))))))))).))).... ( -33.20) >DroEre_CAF1 253283 93 + 1 CUGAAGAUAUUUCUUAAAGCUUU-UCCUC-UGUCGGCGUGCUUUCCCCAUUAGCGCAACUGCUGAGUCACCAGACAACUGGUGGCUAUGUCCCAU .....(((((.............-.....-..((((((((((.........)))))....)))))((((((((....)))))))).))))).... ( -24.80) >DroYak_CAF1 266746 95 + 1 CUGAAAAUAUUUCUUAAAGCUUUUUCCUCUUGUCGGGGUGUUUUCCCCAUUAGCGCGGCUGCUGAGUCACCAGACAUCUGGUGGCUCUGGCCCAU ..((((((((..(...(((.........)))...)..)))))))).........(.((((...((((((((((....)))))))))).))))).. ( -33.10) >consensus CUCAAGAUAUUUCUUAAAGCUUUUUCCUCUUGUCG_GGUGUUUUCCCCAUUAGAGCGGCUGCUGAGUCACCAGACAUCUGGUGGCUCUGGCUCAU ..(((((((((.....(((.........))).....)))))))))...........((((...((((((((((....)))))))))).))))... (-20.38 = -21.54 + 1.16)

| Location | 18,414,105 – 18,414,199 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 86.65 |
| Mean single sequence MFE | -33.18 |
| Consensus MFE | -26.90 |
| Energy contribution | -28.10 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.976880 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18414105 94 + 22407834 UUGUUUUCCCCAUUAGAGCAGCUGCUGAGUCACCAGACAUCUGGUGGCUCUGGCUCAUUUGUCCGGCAGAGUUCUACUUUCC-CCCCUUAGGGCU .............((((((..(((((((((((((((....)))))))))).(((......))).))))).))))))......-(((....))).. ( -34.70) >DroSec_CAF1 254709 95 + 1 GUGUUUUGCCCAUUAGAGCGGCUGCUGAGUCACCAGACAUCUGGUGGCUCUGGCUCAUUUGUCCGGCAGAGUUCCACUUGCCACCCCCCAACGCU (((((..((((......).))).(((((((((((((....)))))))))).)))..........(((((........))))).......))))). ( -34.00) >DroSim_CAF1 266365 95 + 1 GUGUUUUGCCCAUUAGAGCGGCUGCUGAGUCACCAGACAUCUGGUGGCUCUGGCUCAUUUGUCCGGCAGAGUUCCACUUGCCACCCCCCAUGGCU ((((((((((...((((..((((...((((((((((....)))))))))).))))..))))...)))))))...)))..((((.......)))). ( -37.40) >DroEre_CAF1 253318 90 + 1 GUGCUUUCCCCAUUAGCGCAACUGCUGAGUCACCAGACAACUGGUGGCUAUGUCCCAUUUGUCCGGCAGAGUUCUAUUU-UU---UCUCAGGGC- (((((.........)))))..(((((((((((((((....)))))))))(((...))).....)))))).(((((....-..---....)))))- ( -26.90) >DroYak_CAF1 266783 91 + 1 GUGUUUUCCCCAUUAGCGCGGCUGCUGAGUCACCAGACAUCUGGUGGCUCUGGCCCAUUUGUCCGGCAGAGUUCUACUUCCC---UCCCAGGGC- (((((.........)))))((((...((((((((((....)))))))))).)))).....((((((.((.(........).)---).)).))))- ( -32.90) >consensus GUGUUUUCCCCAUUAGAGCGGCUGCUGAGUCACCAGACAUCUGGUGGCUCUGGCUCAUUUGUCCGGCAGAGUUCUACUUGCC_CCCCCCAGGGCU .............((((((..(((((((((((((((....)))))))))).(((......))).))))).))))))................... (-26.90 = -28.10 + 1.20)

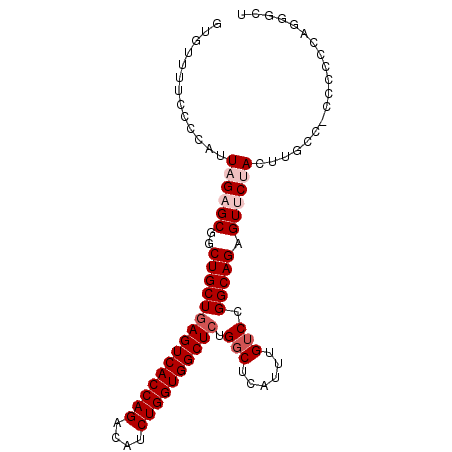
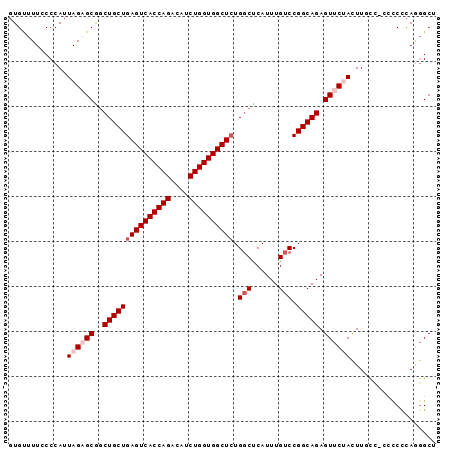
| Location | 18,414,123 – 18,414,238 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 80.15 |
| Mean single sequence MFE | -36.16 |
| Consensus MFE | -20.20 |
| Energy contribution | -21.03 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.931672 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
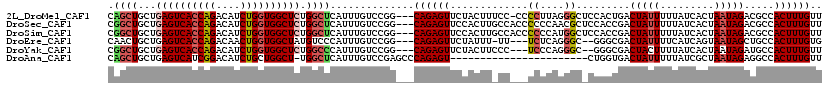
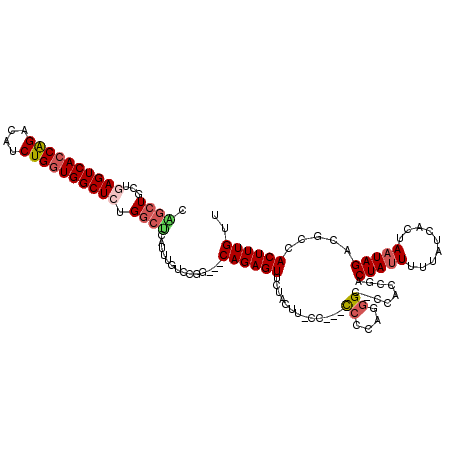
>2L_DroMel_CAF1 18414123 115 + 22407834 CAGCUGCUGAGUCACCAGACAUCUGGUGGCUCUGGCUCAUUUGUCCGG---CAGAGUUCUACUUUCC-CCCCUUAGGGCUCCACUGACUAUUUUUAUCACUAAUAGACGCCACUUUGUU .((((...((((((((((....)))))))))).)))).........((---(.((((((((......-.....))))))))......(((((.........)))))..)))........ ( -36.10) >DroSec_CAF1 254727 116 + 1 CGGCUGCUGAGUCACCAGACAUCUGGUGGCUCUGGCUCAUUUGUCCGG---CAGAGUUCCACUUGCCACCCCCCAACGCUCCACCGACUAUUUUUAUCACUAAUAGACGCCACUUUGUU .(((.(((((((((((((....)))))))))).)))......(((.((---(((........))))).........((......))...................))))))........ ( -35.00) >DroSim_CAF1 266383 116 + 1 CGGCUGCUGAGUCACCAGACAUCUGGUGGCUCUGGCUCAUUUGUCCGG---CAGAGUUCCACUUGCCACCCCCCAUGGCUCCACCGACUAUUUUUAUCACUAAUAGACGCCACUUUGUU .(((.(((((((((((((....)))))))))).)))......(((.((---(((........)))))........(((.....)))...................))))))........ ( -36.50) >DroEre_CAF1 253336 110 + 1 CAACUGCUGAGUCACCAGACAACUGGUGGCUAUGUCCCAUUUGUCCGG---CAGAGUUCUAUUU-UU---UCUCAGGGC--GGGCGACUAUUUUCAUCAGUAAUAGCUGCCACUUUGUG ...(((((((((((((((....)))))))))(((...))).....)))---)))..........-..---...(((((.--.((((.(((((.........))))).)))).))))).. ( -32.00) >DroYak_CAF1 266801 111 + 1 CGGCUGCUGAGUCACCAGACAUCUGGUGGCUCUGGCCCAUUUGUCCGG---CAGAGUUCUACUUCCC---UCCCAGGGC--GGGCGACUACUUUUAUCACUAAUAGAUGCCACUUUGUU .((((...((((((((((....)))))))))).)))).........((---((((((.......(((---.....))).--.((((.(((.((........))))).)))))))))))) ( -39.10) >DroAna_CAF1 233178 95 + 1 CAGCUGCUGAGUCAUCGGACAUCUGCUGGCU-UGGCUCAUUUGUCCGAGCCCAGAGU-----------------------CUGGUGACUAUUUUUAUCGCUAAUAGAGGCCACUUUGUU .(((.....(((((((((((.((((..((((-((((......).)))))))))))))-----------------------))))))))).........)))(((((((....))))))) ( -38.24) >consensus CAGCUGCUGAGUCACCAGACAUCUGGUGGCUCUGGCUCAUUUGUCCGG___CAGAGUUCUACUU_CC___CCCCAGGGC_CCACCGACUAUUUUUAUCACUAAUAGACGCCACUUUGUU .((((...((((((((((....)))))))))).))))..............((((((.............((....)).........(((((.........))))).....)))))).. (-20.20 = -21.03 + 0.83)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:13:42 2006