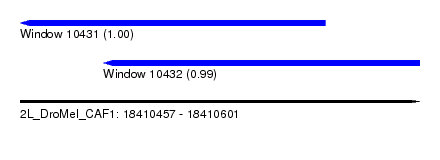
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,410,457 – 18,410,601 |
| Length | 144 |
| Max. P | 0.998078 |

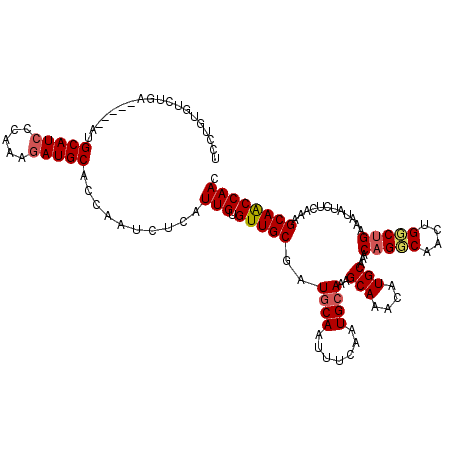
| Location | 18,410,457 – 18,410,567 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 94.91 |
| Mean single sequence MFE | -25.12 |
| Consensus MFE | -20.98 |
| Energy contribution | -21.30 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.84 |
| SVM decision value | 3.00 |
| SVM RNA-class probability | 0.998078 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
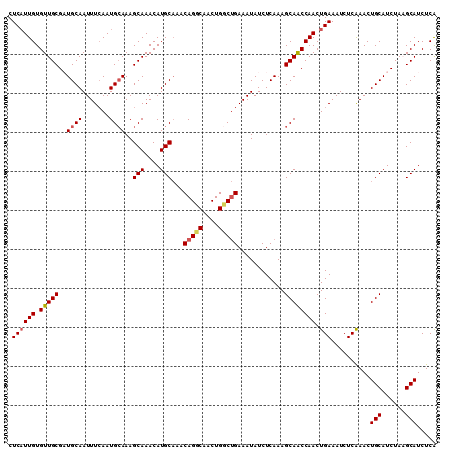
>2L_DroMel_CAF1 18410457 110 - 22407834 CUCAUUGUGUUGCGAUGCAAUUUCAAUGCAAAGCAAACAUGCAAACAGGCAACUGGCUGAAAUAUCUCAAAGCAACCAACUGAAAUCUCAAACUGCAUCUAAGCAUCUCA .((((((.(((((..((..((((((..((...(((....)))...(((....)))))))))))....))..)))))))).)))..........(((......)))..... ( -24.50) >DroSec_CAF1 250730 110 - 1 CUCAUUGUGUUGCGAUGCAAUUUCAAUGCAAAGCAAACAUGCAAACAGCCAACUGGCUGAAAUAUCUCAAAGCAACCAACUGAAAUCUCAAACUGCAUCUAAGCAUCUCA .((((((.(((((..((((.......))))..(((....)))...(((((....)))))............)))))))).)))..........(((......)))..... ( -25.40) >DroSim_CAF1 262393 110 - 1 CUCAUUGUGUUGCGAUGCAAUUUCAAUGCAAAGCAAACAUGCAAACAGCCAACUGGCUGAAAUAUCUCAAAGCAACCAACUGAAAUCUCAAACUGCAUCUAAGCAUCUCA .((((((.(((((..((((.......))))..(((....)))...(((((....)))))............)))))))).)))..........(((......)))..... ( -25.40) >DroEre_CAF1 249440 110 - 1 CUCAUUGUGUUGCGCUGCAAUUUGAAUGCAAAGCAAACAUGCUAACAGGCAACUGCCCGAAAUAUCUCAAAGCAACCAACUGAAAUCUCAAACUGCAUCUACGCAUCUCG .((((((.(((((..((((.......)))).((((....))))..(((....)))................)))))))).)))..........(((......)))..... ( -21.60) >DroYak_CAF1 262397 110 - 1 CUCCUUGUGUUGCGCUGCAAUUUCAAUGAAAAGCAAACAUGCAAACAGGCAACUGCCUGAAAUAUCUCAAAGCAGCCAACUGAAAUCUCGAACUGCAUCUACGCAUCUCA ........((((.(((((..............(((....)))...(((((....)))))............))))))))).........((..(((......)))..)). ( -28.70) >consensus CUCAUUGUGUUGCGAUGCAAUUUCAAUGCAAAGCAAACAUGCAAACAGGCAACUGGCUGAAAUAUCUCAAAGCAACCAACUGAAAUCUCAAACUGCAUCUAAGCAUCUCA .((((((.(((((..((((.......))))..(((....)))...(((((....)))))............)))))))).)))..........(((......)))..... (-20.98 = -21.30 + 0.32)

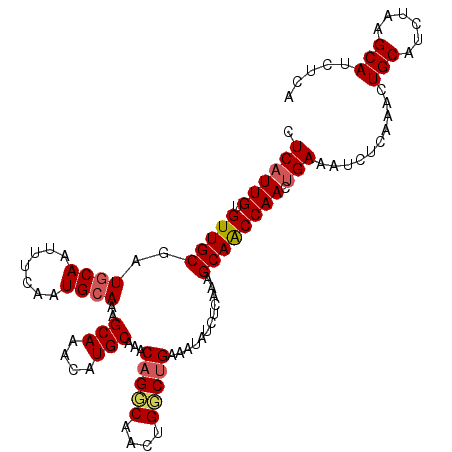
| Location | 18,410,487 – 18,410,601 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.66 |
| Mean single sequence MFE | -30.60 |
| Consensus MFE | -23.38 |
| Energy contribution | -23.70 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.76 |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.989691 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18410487 114 - 22407834 UCCUGUGUCUGA------UGCAUCCCAAAGAUGCAGCAAUCUCAUUGUGUUGCGAUGCAAUUUCAAUGCAAAGCAAACAUGCAAACAGGCAACUGGCUGAAAUAUCUCAAAGCAACCAAC ...((.(..((.------((((((.....)))))).))..).))(((.(((((..((..((((((..((...(((....)))...(((....)))))))))))....))..)))))))). ( -29.90) >DroSec_CAF1 250760 120 - 1 UCCUGUGUCUGAUGCUGAUGCAUGCCAAAGAUGCAGCAAUCUCAUUGUGUUGCGAUGCAAUUUCAAUGCAAAGCAAACAUGCAAACAGCCAACUGGCUGAAAUAUCUCAAAGCAACCAAC ..((((((((.((((....)))).....))))))))........(((.(((((..((((.......))))..(((....)))...(((((....)))))............)))))))). ( -33.40) >DroSim_CAF1 262423 120 - 1 UCCUGUGUCUGAUGCUGAUGCAUCCCAAAGAUGCACCAAUCUCAUUGUGUUGCGAUGCAAUUUCAAUGCAAAGCAAACAUGCAAACAGCCAACUGGCUGAAAUAUCUCAAAGCAACCAAC ...((((((((((((....)))))....))))))).........(((.(((((..((((.......))))..(((....)))...(((((....)))))............)))))))). ( -33.30) >DroEre_CAF1 249470 104 - 1 ----------G------AAGCAUCCCAAAGAUGCACCAUGCUCAUUGUGUUGCGCUGCAAUUUGAAUGCAAAGCAAACAUGCUAACAGGCAACUGCCCGAAAUAUCUCAAAGCAACCAAC ----------.------..(((((.....)))))..........(((.(((((..((((.......)))).((((....))))..(((....)))................)))))))). ( -24.20) >DroYak_CAF1 262427 114 - 1 UCCUGUGCCAG------AAGCAUCCCAAAGAUGCAUCAUGCUCCUUGUGUUGCGCUGCAAUUUCAAUGAAAAGCAAACAUGCAAACAGGCAACUGCCUGAAAUAUCUCAAAGCAGCCAAC ......((..(------(.(((((.....))))).))..)).......((((.(((((..............(((....)))...(((((....)))))............))))))))) ( -32.20) >consensus UCCUGUGUCUGA_____AUGCAUCCCAAAGAUGCACCAAUCUCAUUGUGUUGCGAUGCAAUUUCAAUGCAAAGCAAACAUGCAAACAGGCAACUGGCUGAAAUAUCUCAAAGCAACCAAC ...................(((((.....)))))..........(((.(((((..((((.......))))..(((....)))...(((((....)))))............)))))))). (-23.38 = -23.70 + 0.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:13:38 2006