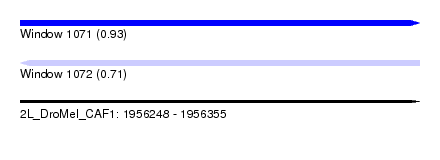
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,956,248 – 1,956,355 |
| Length | 107 |
| Max. P | 0.928392 |

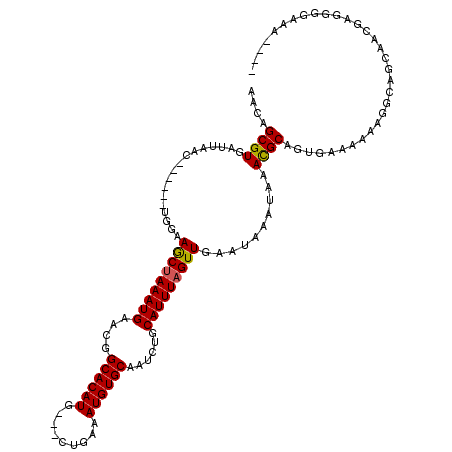
| Location | 1,956,248 – 1,956,355 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.92 |
| Mean single sequence MFE | -23.12 |
| Consensus MFE | -18.51 |
| Energy contribution | -18.39 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928392 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1956248 107 + 22407834 AACAGCGUGAUUAAC------UGGAAGCUAAAUGAACGGCACAUA---CUGAAAUGUGCAAUCUGCAUUUAGUUGAAUAAAUAAACGCAGUGAAAAAAGGCAGCAACGAGGGGAAA---- ....((((.......------....(((((((((....((((((.---.....))))))......)))))))))..........))))............................---- ( -23.10) >DroSim_CAF1 599 107 + 1 AACAGCGUGAUUAAC------UGGAAGCUAAAUGAACGGCACAUA---CUGAAAUGUGCAAUCUGCAUUUAGUUGAAUAAAUAAACGCAGUGAAAAAAGGCAGCAACGAGGGGAAA---- ....((((.......------....(((((((((....((((((.---.....))))))......)))))))))..........))))............................---- ( -23.10) >DroEre_CAF1 7890 111 + 1 AACAGCGUGAUUAAC------UGGAAGCUAAAUGAACGGCACAUG---CUGAAAUGUGCAAUCUGCAUUUAGUUGAAUAAAUAAACGCAGUGAAAAAAGGCAGCAACGAGGGGAAAAUGG .....(((..((..(------(.(.(((((((((....((((((.---.....))))))......)))))))))............((..(....)...)).....).))..))..))). ( -24.40) >DroYak_CAF1 5597 113 + 1 AACAGCGUGAUUAACUACUACUGAAAGCUAAAUGAACGGCACAUG---CUGAAAUGUGCAAUCUGCAUUUAGUUGAAUAAAUAAACGCGGUGAAAAAAGGCAGCGACGAGGGGAAA---- ..(..(((.................(((((((((....((((((.---.....))))))......)))))))))...........(((.((........)).))))))..).....---- ( -24.90) >DroAna_CAF1 28331 98 + 1 AACAGCGUUAUUAAC------UGGAAACGAAAUGACCUGCACAUGAUGCAGAAAUGUGCAAUCUGCAUUUAGUUGAAUAAAUAAAUGCAACAAAAAAA-----CAACGA----------- ...((.((((((...------((....)).))))))))((((((.........))))))....((((((((..........)))))))).........-----......----------- ( -20.10) >consensus AACAGCGUGAUUAAC______UGGAAGCUAAAUGAACGGCACAUG___CUGAAAUGUGCAAUCUGCAUUUAGUUGAAUAAAUAAACGCAGUGAAAAAAGGCAGCAACGAGGGGAAA____ ....((((.................(((((((((....((((((.........))))))......)))))))))..........))))................................ (-18.51 = -18.39 + -0.12)

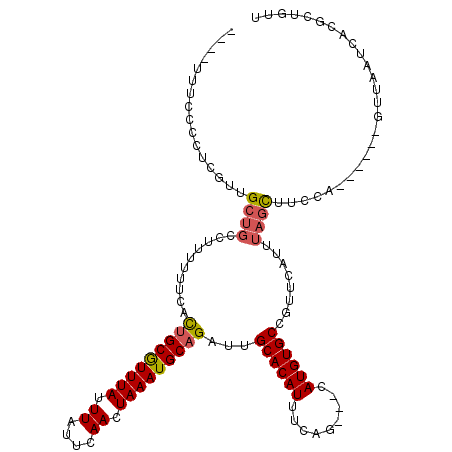
| Location | 1,956,248 – 1,956,355 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.92 |
| Mean single sequence MFE | -20.52 |
| Consensus MFE | -14.90 |
| Energy contribution | -15.22 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.708712 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1956248 107 - 22407834 ----UUUCCCCUCGUUGCUGCCUUUUUUCACUGCGUUUAUUUAUUCAACUAAAUGCAGAUUGCACAUUUCAG---UAUGUGCCGUUCAUUUAGCUUCCA------GUUAAUCACGCUGUU ----............((((..........(((((((((.((....)).)))))))))...((((((.....---.))))))........))))...((------((.......)))).. ( -21.20) >DroSim_CAF1 599 107 - 1 ----UUUCCCCUCGUUGCUGCCUUUUUUCACUGCGUUUAUUUAUUCAACUAAAUGCAGAUUGCACAUUUCAG---UAUGUGCCGUUCAUUUAGCUUCCA------GUUAAUCACGCUGUU ----............((((..........(((((((((.((....)).)))))))))...((((((.....---.))))))........))))...((------((.......)))).. ( -21.20) >DroEre_CAF1 7890 111 - 1 CCAUUUUCCCCUCGUUGCUGCCUUUUUUCACUGCGUUUAUUUAUUCAACUAAAUGCAGAUUGCACAUUUCAG---CAUGUGCCGUUCAUUUAGCUUCCA------GUUAAUCACGCUGUU ................((((..........(((((((((.((....)).)))))))))...((((((.....---.))))))........))))...((------((.......)))).. ( -20.50) >DroYak_CAF1 5597 113 - 1 ----UUUCCCCUCGUCGCUGCCUUUUUUCACCGCGUUUAUUUAUUCAACUAAAUGCAGAUUGCACAUUUCAG---CAUGUGCCGUUCAUUUAGCUUUCAGUAGUAGUUAAUCACGCUGUU ----............((((............(((((((.((....)).))))))).((..((((((.....---.))))))...))...))))...((((.((........)))))).. ( -18.30) >DroAna_CAF1 28331 98 - 1 -----------UCGUUG-----UUUUUUUGUUGCAUUUAUUUAUUCAACUAAAUGCAGAUUGCACAUUUCUGCAUCAUGUGCAGGUCAUUUCGUUUCCA------GUUAAUAACGCUGUU -----------.(((((-----((...(((.((((((((.((....)).))))))))((((((((((.........))))))).)))..........))------)..)))))))..... ( -21.40) >consensus ____UUUCCCCUCGUUGCUGCCUUUUUUCACUGCGUUUAUUUAUUCAACUAAAUGCAGAUUGCACAUUUCAG___CAUGUGCCGUUCAUUUAGCUUCCA______GUUAAUCACGCUGUU ................((((..........(((((((((.((....)).)))))))))...((((((.........))))))........)))).......................... (-14.90 = -15.22 + 0.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:42:05 2006