
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,382,211 – 18,382,385 |
| Length | 174 |
| Max. P | 0.999350 |

| Location | 18,382,211 – 18,382,327 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.44 |
| Mean single sequence MFE | -48.55 |
| Consensus MFE | -28.88 |
| Energy contribution | -28.99 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.28 |
| Mean z-score | -3.29 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.781092 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
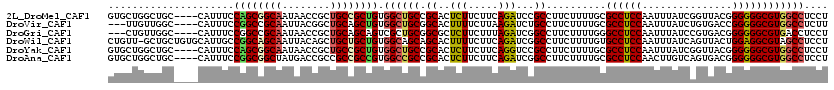
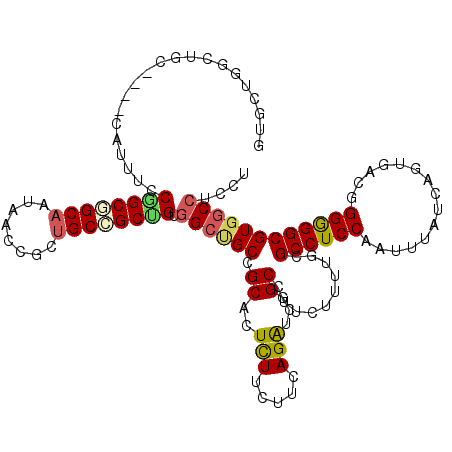
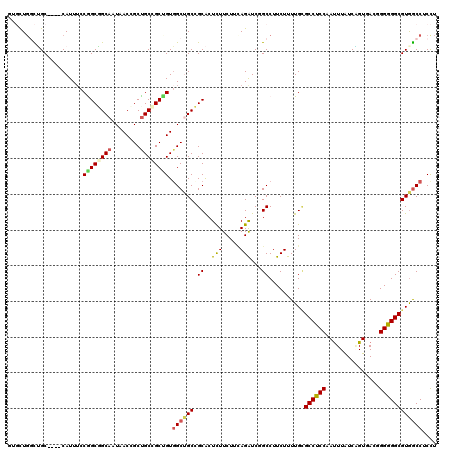
>2L_DroMel_CAF1 18382211 116 - 22407834 GUGCUGGCUGC----CAUUUCCAGCGGCAAUAACCGCUGCCGCUGUGGCUGCCGCACUCUUCUUCAGAUCCGCCUUCUUUUGCGCCUCCAAUUUAUCGGUUACGGGGGGCGUGGCCUCCU ((((.(((.((----((....((((((((........)))))))))))).)))))))..............(((.......((((((((.......((....)))))))))))))..... ( -48.02) >DroVir_CAF1 341039 113 - 1 ---UUGUUGGC----CAUUUCCGGCCGCAAUUACGGCUGCAGCUGUGGCUGCGGCACUUUUCUUAAGAUCUGCCUUCUUUUGCGCCUCCAAUUUAUCUGUGACCGGGGGCGUGGCCUCUU ---.....(((----(.......((((((.(((((((....))))))).))))))..........................((((((((...............)))))))))))).... ( -43.06) >DroGri_CAF1 274533 113 - 1 ---CUGUUGGC----CAUUUCCGGCCGCAAUAACCGCUGCAGCAGUCGCUGCGGCGCUCUUCUUUAGAUCGGCCUUCUUUUGGGCCUCCAAUUUAUCCGUGACGGGGGGCGUGACCUCCU ---..((((((----(......))))((......(((((((((....)))))))))..............(((((......)))))............)))))(((((......))))). ( -43.80) >DroWil_CAF1 300264 119 - 1 CUGUU-GCUGCUGUGCAUUGCCGGCAGCAAUUACAGCUGCUGCUGUGGCAGCAGCACUUUUCUUCAGAUCGGCCUUCUUUUGUGCCUCCAAUUUAUCAGUUACUGGAGGCGUAGCCUCCU (((((-(((((((.((...))))))))))).....((((((((....)))))))).........)))...(((.......((((((((((.............))))))))))))).... ( -47.63) >DroYak_CAF1 236329 116 - 1 GUGCUGGCUGC----CAUUUCCAGCGGCAAUAACCGCUGCCGCUGUGGCUGCCGCACUCUUCUUCAGGUCCGCCUUCUUUUGCGCCUCCAAUUUAUCGGUUACGGGGGGCGUGGCCUCCU ((((.(((.((----((....((((((((........)))))))))))).)))))))........(((((((((((((.....(((...........)))...)))))))).)))))... ( -52.20) >DroAna_CAF1 205708 116 - 1 GUGCUGGCUGC----CAUUUCCGGCGGCUAUGACCGCCGCCGCCGUGGCCGCCGCACUCUUCUUCAGAUCGGCCUUCUUUUGCGCCUCCAACUUGUCAGUGACGGGGGGCGUGGCCUCCU ((((.(((.((----(((...(((((((.......)))))))..))))).))))))).............((((.......((((((((.(((....)))....)))))))))))).... ( -56.61) >consensus GUGCUGGCUGC____CAUUUCCGGCGGCAAUAACCGCUGCCGCUGUGGCUGCCGCACUCUUCUUCAGAUCGGCCUUCUUUUGCGCCUCCAAUUUAUCAGUGACGGGGGGCGUGGCCUCCU .....................((((((((........)))))))).((((((.((..(((.....)))...))..........((((((...............)))))))))))).... (-28.88 = -28.99 + 0.11)
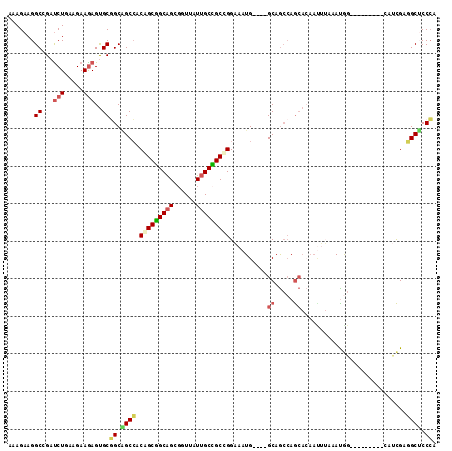
| Location | 18,382,251 – 18,382,352 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 77.64 |
| Mean single sequence MFE | -39.20 |
| Consensus MFE | -21.60 |
| Energy contribution | -21.85 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.32 |
| Mean z-score | -3.14 |
| Structure conservation index | 0.55 |
| SVM decision value | 2.30 |
| SVM RNA-class probability | 0.991963 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
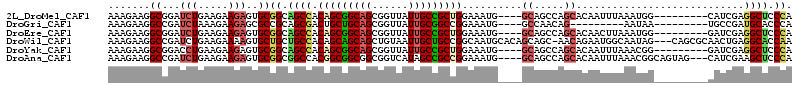
>2L_DroMel_CAF1 18382251 101 + 22407834 AAAGAAGGCGGAUCUGAAGAAGAGUGCGGCAGCCACAGCGGCAGCGGUUAUUGCCGCUGGAAAUG----GCAGCCAGCACAAUUUAAAUGG---------CAUCGAGGCUCCCA ...(..(((...(((.....)))(((((((.(((((((((((((......)))))))))....))----)).))).))))...........---------.......)))..). ( -36.60) >DroGri_CAF1 274573 92 + 1 AAAGAAGGCCGAUCUAAAGAAGAGCGCCGCAGCGACUGCUGCAGCGGUUAUUGCGGCCGGAAAUG----GCCAACAG---------AAUAA---------UGCCGAUGCACCCA ......(((((.(((.....)))(.(((((((..(((((....)))))..)))))))).....))----))).....---------.....---------(((....))).... ( -32.40) >DroEre_CAF1 226533 101 + 1 AAAGAAGGCGGAUCUGAAGAAGAGUGCGGCAGCCACAGCGGCAGCGGUUAUUGCCGCUGGAAAUG----GCAGCCAGCACAACUUAAAUGG---------GAUCGAGGCUCCCA .....((.(.(((((.(..(((.(((((((.(((((((((((((......)))))))))....))----)).))).))))..)))...).)---------))))..).)).... ( -40.30) >DroWil_CAF1 300304 110 + 1 AAAGAAGGCCGAUCUGAAGAAAAGUGCUGCUGCCACAGCAGCAGCUGUAAUUGCUGCCGGCAAUGCACAGCAGC-AACAGAAUGGCAAUAG---CAGCGCAACUGAGGCACCAA .......(((..((....))...((((((((((((((((....)))))..(((((((.(((...)).).)))))-))......)))...))---))))))......)))..... ( -41.10) >DroYak_CAF1 236369 101 + 1 AAAGAAGGCGGACCUGAAGAAGAGUGCGGCAGCCACAGCGGCAGCGGUUAUUGCCGCUGGAAAUG----GCAGCCAGCACAAUUUAAACGG---------GAUCGAGGCUCCCA ..((....(((.((((..(((..(((((((.(((((((((((((......)))))))))....))----)).))).))))..)))...)))---------).)))...)).... ( -40.20) >DroAna_CAF1 205748 107 + 1 AAAGAAGGCCGAUCUGAAGAAGAGUGCGGCGGCCACGGCGGCGGCGGUCAUAGCCGCCGGAAAUG----GCAGCCAGCACAAUUUAAACGGCAGUAG---CAUCGAAGCUCCCA .......((((.(((.....)))(((((((.((((...(((((((.......)))))))....))----)).))).))))........)))).(.((---(......))).).. ( -44.60) >consensus AAAGAAGGCCGAUCUGAAGAAGAGUGCGGCAGCCACAGCGGCAGCGGUUAUUGCCGCCGGAAAUG____GCAGCCAGCACAAUUUAAAUGG_________CAUCGAGGCUCCCA .......((...(((.....)))..))((.((((.(((((((((......)))))))))..........((.....))............................)))).)). (-21.60 = -21.85 + 0.25)

| Location | 18,382,251 – 18,382,352 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 77.64 |
| Mean single sequence MFE | -39.52 |
| Consensus MFE | -19.19 |
| Energy contribution | -19.12 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.43 |
| Mean z-score | -3.32 |
| Structure conservation index | 0.49 |
| SVM decision value | 2.35 |
| SVM RNA-class probability | 0.992764 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18382251 101 - 22407834 UGGGAGCCUCGAUG---------CCAUUUAAAUUGUGCUGGCUGC----CAUUUCCAGCGGCAAUAACCGCUGCCGCUGUGGCUGCCGCACUCUUCUUCAGAUCCGCCUUCUUU ..(((((......)---------)..........((((.(((.((----((....((((((((........)))))))))))).)))))))...........)))......... ( -35.40) >DroGri_CAF1 274573 92 - 1 UGGGUGCAUCGGCA---------UUAUU---------CUGUUGGC----CAUUUCCGGCCGCAAUAACCGCUGCAGCAGUCGCUGCGGCGCUCUUCUUUAGAUCGGCCUUCUUU .((.(((...((((---------.....---------.))))(((----(......)))))))....))((((((((....))))))))(((..((....))..)))....... ( -33.60) >DroEre_CAF1 226533 101 - 1 UGGGAGCCUCGAUC---------CCAUUUAAGUUGUGCUGGCUGC----CAUUUCCAGCGGCAAUAACCGCUGCCGCUGUGGCUGCCGCACUCUUCUUCAGAUCCGCCUUCUUU .(((((.(..((((---------......(((..((((.(((.((----((....((((((((........)))))))))))).))))))).))).....)))).).))))).. ( -39.40) >DroWil_CAF1 300304 110 - 1 UUGGUGCCUCAGUUGCGCUG---CUAUUGCCAUUCUGUU-GCUGCUGUGCAUUGCCGGCAGCAAUUACAGCUGCUGCUGUGGCAGCAGCACUUUUCUUCAGAUCGGCCUUCUUU ..(((((.......))))).---.....(((..((((((-(((((((.((...))))))))))).....((((((((....)))))))).........))))..)))....... ( -45.30) >DroYak_CAF1 236369 101 - 1 UGGGAGCCUCGAUC---------CCGUUUAAAUUGUGCUGGCUGC----CAUUUCCAGCGGCAAUAACCGCUGCCGCUGUGGCUGCCGCACUCUUCUUCAGGUCCGCCUUCUUU .((((.......))---------)).........((((.(((.((----((....((((((((........)))))))))))).)))))))........(((....)))..... ( -40.00) >DroAna_CAF1 205748 107 - 1 UGGGAGCUUCGAUG---CUACUGCCGUUUAAAUUGUGCUGGCUGC----CAUUUCCGGCGGCUAUGACCGCCGCCGCCGUGGCCGCCGCACUCUUCUUCAGAUCGGCCUUCUUU .((.(((......)---)).))((((........((((.(((.((----(((...(((((((.......)))))))..))))).)))))))(((.....))).))))....... ( -43.40) >consensus UGGGAGCCUCGAUC_________CCAUUUAAAUUGUGCUGGCUGC____CAUUUCCAGCGGCAAUAACCGCUGCCGCUGUGGCUGCCGCACUCUUCUUCAGAUCCGCCUUCUUU .((.((((............................((.....))..........((((((((........)))))))).)))).))((..(((.....)))...))....... (-19.19 = -19.12 + -0.08)

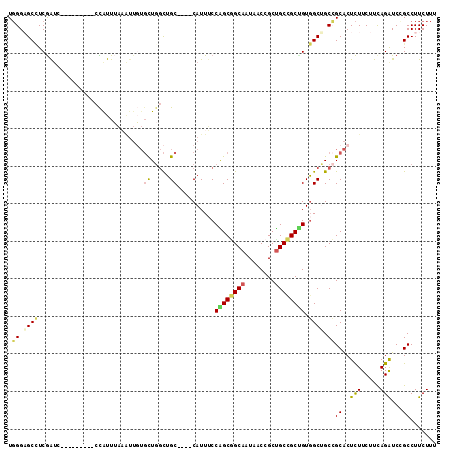
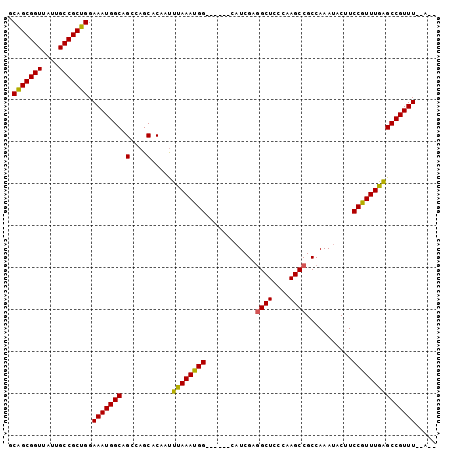
| Location | 18,382,291 – 18,382,385 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 91.68 |
| Mean single sequence MFE | -32.89 |
| Consensus MFE | -31.62 |
| Energy contribution | -30.93 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.96 |
| SVM decision value | 3.53 |
| SVM RNA-class probability | 0.999350 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18382291 94 + 22407834 GCAGCGGUUAUUGCCGCUGGAAAUGGCAGCCAGCACAAUUUAAAUGG------CAUCGAGGCUCCCAAGCCGCCAAAUACUUCCGUUUAAGCCGUUU--AGG .(((((((....))))))).(((((((.((((............)))------).....((((....))))...................)))))))--... ( -32.10) >DroSec_CAF1 226040 92 + 1 GCAGCGGUUAUUGCCGCUGGAAAUGGCAGCCAGCACAAUUUAAAUGG------CAUCGAGGCUCCCAAGCCGCCAAAUACUUCCGUUUGAGCCGUUU--A-- .(((((((....))))))).(((((((.((((............)))------).....((((....))))..(((((......))))).)))))))--.-- ( -33.30) >DroSim_CAF1 234620 92 + 1 GCAGCGGUUAUUGCCGCUGGAAAUGGCAGCCAGCACAAUUUAAAUGG------CAUCGAGGCUCCCAAGCCGCCAAAUACUUCCGUUUGAGCCGUUU--A-- .(((((((....))))))).(((((((.((((............)))------).....((((....))))..(((((......))))).)))))))--.-- ( -33.30) >DroEre_CAF1 226573 92 + 1 GCAGCGGUUAUUGCCGCUGGAAAUGGCAGCCAGCACAACUUAAAUGG------GAUCGAGGCUCCCAAGCCGCCAAAUACUUCCGUUUGAGCCGUUU--A-- .(((((((....))))))).(((((((.(....).....((((((((------((....((((....)))).........)))))))))))))))))--.-- ( -32.32) >DroYak_CAF1 236409 92 + 1 GCAGCGGUUAUUGCCGCUGGAAAUGGCAGCCAGCACAAUUUAAACGG------GAUCGAGGCUCCCAAGCCGCCAAAUACUUCCGUUUGAGCCGUUU--A-- .(((((((....))))))).(((((((.(....).....((((((((------((....((((....)))).........)))))))))))))))))--.-- ( -34.22) >DroAna_CAF1 205788 100 + 1 GCGGCGGUCAUAGCCGCCGGAAAUGGCAGCCAGCACAAUUUAAACGGCAGUAGCAUCGAAGCUCCCAAGCCGCCAAACACGUCCGUUUAGGCCGUUUUGA-- .(((((((....)))))))((((((((.................((((.(.(((......))).)...))))..((((......))))..))))))))..-- ( -32.10) >consensus GCAGCGGUUAUUGCCGCUGGAAAUGGCAGCCAGCACAAUUUAAAUGG______CAUCGAGGCUCCCAAGCCGCCAAAUACUUCCGUUUGAGCCGUUU__A__ .(((((((....))))))).(((((((.(....).....((((((((............((((....))))...........)))))))))))))))..... (-31.62 = -30.93 + -0.69)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:13:30 2006