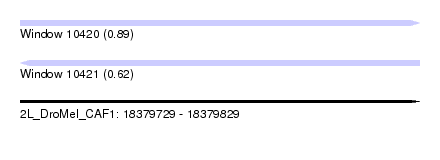
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,379,729 – 18,379,829 |
| Length | 100 |
| Max. P | 0.890574 |

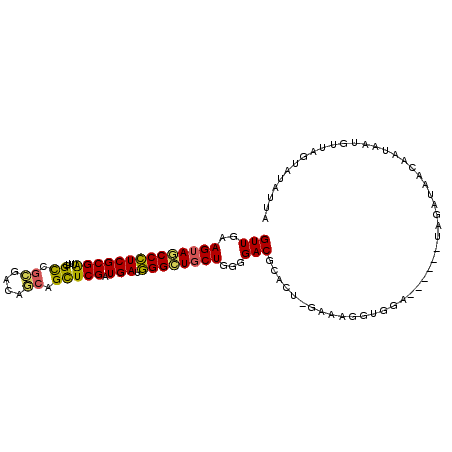
| Location | 18,379,729 – 18,379,829 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 80.13 |
| Mean single sequence MFE | -24.22 |
| Consensus MFE | -16.07 |
| Energy contribution | -18.02 |
| Covariance contribution | 1.95 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.890574 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18379729 100 + 22407834 UAAAAUACUAACAUAAUUGUUAUCUA------UCCACCUUUC-AGUGCGUCCCCAGCAGCCCAUCAUCGAGCUGUUGUCGCGGCCAAAUCGCGAGGGCUACUUCAAC ........(((((....)))))....------..........-((((.((((.(((((((.(......).)))))))((((((.....))))))))))))))..... ( -26.00) >DroSec_CAF1 223513 100 + 1 UAAUAUACUAACUUUAUUGUUAUCCA------UCCAACUUUC-AGUGCGUCCCCAGCAGCCCAUCAUCGAGCUGCUGUCGCGGCCAAAUCGCGAGGGCUACUUCAAC ........((((......))))....------..........-((((.((((.(((((((.(......).)))))))((((((.....))))))))))))))..... ( -27.40) >DroSim_CAF1 232096 100 + 1 UAAUAUACUAACUUUAUUGUUAUCCA------UCCAACUUUC-AGUGCGUCCCCAGCAGCCCAUCAUCGAGCUGCUGUCGCGGCCAAAUCGCGAGGGAUACUUCAAC ........((((......))))....------..........-.....((((((((((((.(......).)))))))((((((.....)))))))))))........ ( -27.50) >DroEre_CAF1 224005 98 + 1 UA-------AACAUUAUUUUC-UCUAUUUGCUCUCACCUUUC-AGUGCGUCCGCAGCAGCCCAUCAUCGAGCUGCUGUCGCGGCCAAAUCGCGAGGGCUACUUCAAC ..-------............-.......((((((.......-..((((...((((((((.(......).))))))))))))((......))))))))......... ( -28.20) >DroYak_CAF1 233920 100 + 1 CAAAAUGCUAACAUUAUUGUCAUCUA------CUCACCUUUC-AGUGCGUCCGCAGCAGCCCAUCAUCGAGCUGCUGUCGCGGCCAAAUCGCGAGGGCUACUUCAAC (((((((....)))).))).......------..........-((((.((((.(((((((.(......).)))))))((((((.....))))))))))))))..... ( -27.30) >DroAna_CAF1 203608 93 + 1 UAAU-------CAUAACACU-CUCCU------CCUGCCUUUCUAGUGCGUCCCCAGCAACCAAUCAUCGAACUAAUGACCAAACCCAACCGCGAUGGUUUCUUCAAC ....-------.........-.....------.............(((.......)))...((((((((......((........))....))))))))........ ( -8.90) >consensus UAAUAUACUAACAUUAUUGUUAUCCA______CCCACCUUUC_AGUGCGUCCCCAGCAGCCCAUCAUCGAGCUGCUGUCGCGGCCAAAUCGCGAGGGCUACUUCAAC ...........................................((((.((((.(((((((.(......).)))))))((((((.....))))))))))))))..... (-16.07 = -18.02 + 1.95)

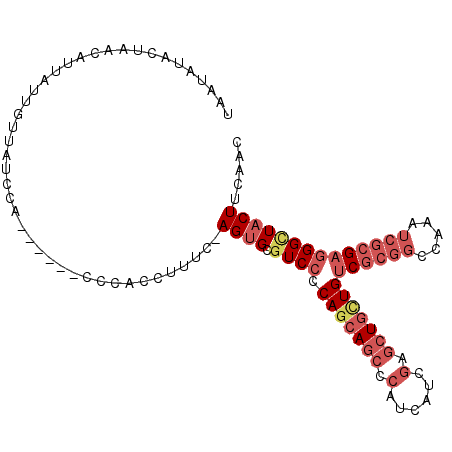
| Location | 18,379,729 – 18,379,829 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 80.13 |
| Mean single sequence MFE | -30.20 |
| Consensus MFE | -20.05 |
| Energy contribution | -19.58 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.618476 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18379729 100 - 22407834 GUUGAAGUAGCCCUCGCGAUUUGGCCGCGACAACAGCUCGAUGAUGGGCUGCUGGGGACGCACU-GAAAGGUGGA------UAGAUAACAAUUAUGUUAGUAUUUUA (((..((((((((((((((.(((..........))).))).))).))))))))...))).((((-....))))((------((..(((((....))))).))))... ( -30.50) >DroSec_CAF1 223513 100 - 1 GUUGAAGUAGCCCUCGCGAUUUGGCCGCGACAGCAGCUCGAUGAUGGGCUGCUGGGGACGCACU-GAAAGUUGGA------UGGAUAACAAUAAAGUUAGUAUAUUA ......((..((((((((.......)))))(((((((((......))))))))))))..))(((-((..((((..------.......))))....)))))...... ( -33.70) >DroSim_CAF1 232096 100 - 1 GUUGAAGUAUCCCUCGCGAUUUGGCCGCGACAGCAGCUCGAUGAUGGGCUGCUGGGGACGCACU-GAAAGUUGGA------UGGAUAACAAUAAAGUUAGUAUAUUA ......(..(((((((((.......)))))(((((((((......)))))))))))))..)(((-((..((((..------.......))))....)))))...... ( -35.60) >DroEre_CAF1 224005 98 - 1 GUUGAAGUAGCCCUCGCGAUUUGGCCGCGACAGCAGCUCGAUGAUGGGCUGCUGCGGACGCACU-GAAAGGUGAGAGCAAAUAGA-GAAAAUAAUGUU-------UA .........((.(((((..((..(..(((.(((((((((......)))))))))....))).).-.))..))))).)).......-............-------.. ( -31.30) >DroYak_CAF1 233920 100 - 1 GUUGAAGUAGCCCUCGCGAUUUGGCCGCGACAGCAGCUCGAUGAUGGGCUGCUGCGGACGCACU-GAAAGGUGAG------UAGAUGACAAUAAUGUUAGCAUUUUG ...(((((.(((((((((.......)))))(((((((((......))))))))).))((.((((-....)))).)------)...(((((....)))))))))))). ( -34.00) >DroAna_CAF1 203608 93 - 1 GUUGAAGAAACCAUCGCGGUUGGGUUUGGUCAUUAGUUCGAUGAUUGGUUGCUGGGGACGCACUAGAAAGGCAGG------AGGAG-AGUGUUAUG-------AUUA ..........((.((.((((.....(..((((((.....))))))..)..)))).))..((.((....)))).))------.....-.........-------.... ( -16.10) >consensus GUUGAAGUAGCCCUCGCGAUUUGGCCGCGACAGCAGCUCGAUGAUGGGCUGCUGGGGACGCACU_GAAAGGUGGA______UAGAUAACAAUAAUGUUAGUAUAUUA (((..((((((((((((((....((.((....)).))))).))).))))))))...)))................................................ (-20.05 = -19.58 + -0.47)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:13:27 2006