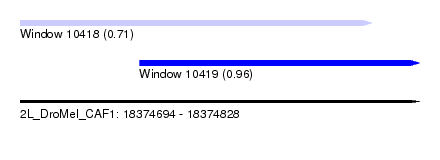
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,374,694 – 18,374,828 |
| Length | 134 |
| Max. P | 0.957237 |

| Location | 18,374,694 – 18,374,812 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 83.86 |
| Mean single sequence MFE | -27.83 |
| Consensus MFE | -20.08 |
| Energy contribution | -20.67 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.708156 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18374694 118 + 22407834 UAUUCACACUCCUGGAGAUAAUCACUUCUGCACCGGAGUUAUCCUAACCAACCGCCAUGUGCUUACUUCAGCACACUGCAUUACAGAGUGGGUAUAUCAUAUAAUUCUAGUUCAUUAC- ((((((((((((((.(((........))).))..)))))..............((..((((((......))))))..))........)))))))........................- ( -26.30) >DroSec_CAF1 218516 118 + 1 UAUUCACACUCCUGGAGAUAAUCACUUCUGCACCGGAGUUAUCCUAACCAAUCGCCAUGUGCUUACUUCAGCACACUGCAUUACAGAGUGGGUAUAUCAUAUGUUUAUUAUUCAUUAC- ((((((((((((((.(((........))).))..)))))..............((..((((((......))))))..))........)))))))........................- ( -26.30) >DroSim_CAF1 227067 118 + 1 UAUUCACACUCCUGGAGAUAAUCACUUCUGCACCGGAGUUAUCCUAACCAAUCGCCAUGUGCUUACUUCAGCACACUGCAUUACAGAGUGGGUAUAUCAUAUGUUUAUUAUUCAUUAC- ((((((((((((((.(((........))).))..)))))..............((..((((((......))))))..))........)))))))........................- ( -26.30) >DroEre_CAF1 219089 117 + 1 UAUUCACUCUCCUGGAGAUAAUCACUUCUGCACCGGAGUUAUCCUAACCAAUCGCCAUGUGCUUACCUCAGCACACUGCAUUACGGAGUGGGUAUA--AUACAUAUCUAAUACAUUAUU ((((((((((..(((.((((...(((((......)))))))))))).......((..((((((......))))))..)).....))))))))))..--..................... ( -27.90) >DroYak_CAF1 228983 119 + 1 UAUCCACUCUCCUGGAGAUAAUCACUUCUGCACCGGAGUUAUCCUUACCAAUCGCCAUGUGCUUACUUCAGCACACUGCAUUACGGAGUGGGUAUAUCAUACAUAUCUAAUUCGUUACU ((((((((((..(((.((..((.(((((......))))).))..)).)))...((..((((((......))))))..)).....))))))))))......................... ( -30.50) >DroAna_CAF1 199288 115 + 1 CGUCCACCAAAAGGGGGAUAACCACUUCUGCACCGGUGUCCUCCUUACAAACCGCCAUAUCCUGACCGCGGCCCACUGCGUUACCGAGUUGGUAUAU--UCAGUUUAAAGCCCA-AAC- ..........((((((((((.((...........)))))))))))).......((..((..((((.(((((....))))).((((.....))))...--))))..))..))...-...- ( -29.70) >consensus UAUUCACACUCCUGGAGAUAAUCACUUCUGCACCGGAGUUAUCCUAACCAAUCGCCAUGUGCUUACUUCAGCACACUGCAUUACAGAGUGGGUAUAUCAUAUAUUUAUAAUUCAUUAC_ (((((((.((((((.(((........))).))..))))...............((..((((((......))))))..))........)))))))......................... (-20.08 = -20.67 + 0.59)

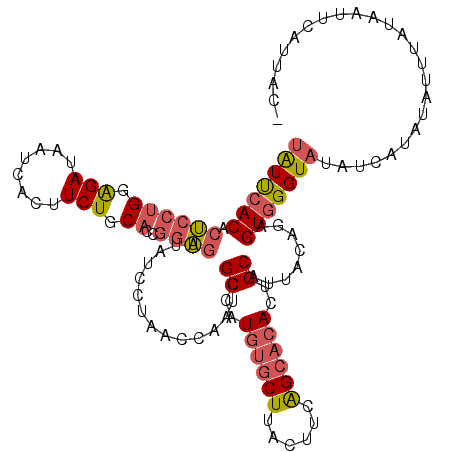
| Location | 18,374,734 – 18,374,828 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 73.93 |
| Mean single sequence MFE | -19.30 |
| Consensus MFE | -10.30 |
| Energy contribution | -9.66 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.957237 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
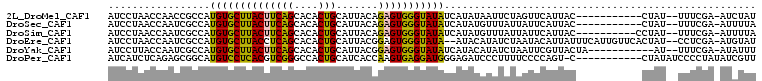
>2L_DroMel_CAF1 18374734 94 + 22407834 AUCCUAACCAACCGCCAUGUGCUUACUUCAGCACACUGCAUUACAGAGUGGGUAUAUCAUAUAAUUCUAGUUCAUUAC-----------CUAU--UUUCGA-AUCUAU .............((..((((((......))))))..)).....((((((((((.....................)))-----------))))--)))...-...... ( -17.70) >DroSec_CAF1 218556 94 + 1 AUCCUAACCAAUCGCCAUGUGCUUACUUCAGCACACUGCAUUACAGAGUGGGUAUAUCAUAUGUUUAUUAUUCAUUAC-----------CUAU--UUUCGA-AUUUUA ...........((((..((((((......))))))..)).....((((((((((....(((.......)))....)))-----------))))--))).))-...... ( -18.90) >DroSim_CAF1 227107 95 + 1 AUCCUAACCAAUCGCCAUGUGCUUACUUCAGCACACUGCAUUACAGAGUGGGUAUAUCAUAUGUUUAUUAUUCAUUAC----------CCUAU--UUUCGA-AUUUUA ...........((((..((((((......))))))..))......(((.(((((....(((.......)))....)))----------))...--.)))))-...... ( -17.60) >DroEre_CAF1 219129 103 + 1 AUCCUAACCAAUCGCCAUGUGCUUACCUCAGCACACUGCAUUACGGAGUGGGUAUA--AUACAUAUCUAAUACAUUAUUUCAUUGUUCACUAU--CCUCGA-AUGUAU .............((..((((((......))))))..))..((((..(.(((((..--..(((.....((((...))))....)))....)))--)).)..-.)))). ( -17.90) >DroYak_CAF1 229023 94 + 1 AUCCUUACCAAUCGCCAUGUGCUUACUUCAGCACACUGCAUUACGGAGUGGGUAUAUCAUACAUAUCUAAUUCGUUACUA-----------AU--UUUCGA-AUAUUU ...........((((..((((((......))))))..))...(((((.(((((((.......))))))).))))).....-----------..--....))-...... ( -18.90) >DroPer_CAF1 266385 96 + 1 AUCAUCUCAGAGCGGCAUGUCCUCACGUCGGGCCACUGCAUCACCAAGUGAGGAUGGGAGAUCCCUUUUCCCCAGU-C-----------CUAUAUCCCCUAUAUCGUU ..........((.(((..((((((((...((............))..))))))))((((((.....))))))..))-)-----------))................. ( -24.80) >consensus AUCCUAACCAAUCGCCAUGUGCUUACUUCAGCACACUGCAUUACAGAGUGGGUAUAUCAUAUAUUUCUAAUUCAUUAC___________CUAU__UUUCGA_AUCUUU .................((((((((((((((....))).......))))))))))).................................................... (-10.30 = -9.66 + -0.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:13:25 2006