
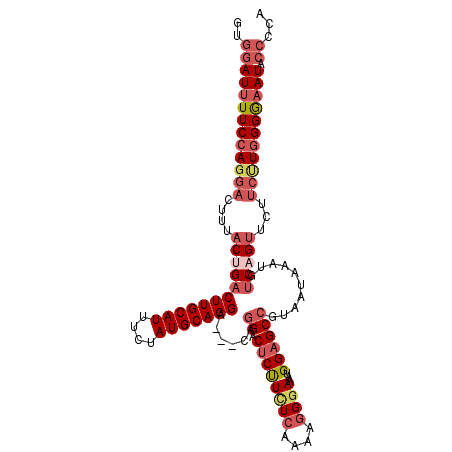
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,367,424 – 18,367,714 |
| Length | 290 |
| Max. P | 0.926558 |

| Location | 18,367,424 – 18,367,532 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 87.34 |
| Mean single sequence MFE | -34.45 |
| Consensus MFE | -26.26 |
| Energy contribution | -28.15 |
| Covariance contribution | 1.89 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.647890 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18367424 108 - 22407834 GUGGAUUUUCCAGGACUUUACUGACUUGCAUUUCUAUGCAGGA---CAGGACUCUUCUCAAAAGGGAAAUGGAGCCGUAAUAAAUGUCAGUUCUUCUUGGGGAAUACCCCA (.(((((..((((((....((((((((((((....))))))).---..((.((((((((....))))...))))))..........)))))...))))))..))).)).). ( -37.40) >DroSec_CAF1 211261 108 - 1 GUGGAUUUUCCAGGACUUUACUGACUUGCAUUUCUAUGCAGGA---CAGGACUCUUCUCAAAAGGGAAAUGGAGCCAUAAUAAAUGUCAGUUCUUCUUGGGGAAUACCCCA (.(((((..((((((....((((((((((((....))))))).---..((.((((((((....))))...))))))..........)))))...))))))..))).)).). ( -37.40) >DroSim_CAF1 220319 108 - 1 GUGGAUUUUCCAGGACUUUACUGACUUGCAUUUCUAUGCAGGA---CAGGACUCUUCUCAAAAGGGAAAUGGAGCCGUAAUAAAUGUCAGUUCUUCUUGGGGAAUACCCCA (.(((((..((((((....((((((((((((....))))))).---..((.((((((((....))))...))))))..........)))))...))))))..))).)).). ( -37.40) >DroEre_CAF1 211718 108 - 1 GUGGAUUUUCCACGACUUUACCGACUUGCAUUUCUAUGCAGGG---CAGGACUCUUUUCAAAAGGGAAAUGGAGCCGUAAUAAAUGUCAGUUCUUCUUAGGGAAUACCCCA ((((.....))))(((.((((...(((((((....))))))).---..((.((((((((......)))).)))))))))).....)))...........(((.....))). ( -31.20) >DroYak_CAF1 221671 108 - 1 GUGGAUUUUCCAGGACUUUACCGACUUGCAUUUCUAUGCAGGG---CAGGACUCUUUUCAAGAGGGAAAUGGAGCCGUAAUAAAUGUCAGUUCUUCCUGGGGAAUACCCCA (.(((((..((((((....((.(((((((((....))))))).---..((.((((((((......)))).))))))..........)).))...))))))..))).)).). ( -35.00) >DroAna_CAF1 192572 102 - 1 GAGGAUUUUCCAGGAAUUUACUGACUUGCAUUUCUAUGCAGGAGGGCGUGCCUCCUGU----AGGAAUACGGUGCCAUAAUAAAUACCAGAUCCAGCUGGGACAUA----- ..(((((((((((((((.((......)).)))))).(((((((((.....))))))))----)))))...((((..........)))).)))))............----- ( -28.30) >consensus GUGGAUUUUCCAGGACUUUACUGACUUGCAUUUCUAUGCAGGA___CAGGACUCUUCUCAAAAGGGAAAUGGAGCCGUAAUAAAUGUCAGUUCUUCUUGGGGAAUACCCCA ..(((((((((((((....((((((((((((....)))))))......((.((((((((....))))...))))))..........)))))...))))))))))).))... (-26.26 = -28.15 + 1.89)


| Location | 18,367,498 – 18,367,612 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 91.23 |
| Mean single sequence MFE | -32.15 |
| Consensus MFE | -27.88 |
| Energy contribution | -27.63 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.620377 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18367498 114 + 22407834 GAAAUGCAAGUCAGUAAAGUCCUGGAAAAUCCACAAGGAUUUGCUCGGUGAUAGCUGAAAGCUUUCUCCCAUUCCCCACAUCGAGGUUUGGAUUUCCCUUUCGGGGAUUUUCCA ((((.((...(((((...(((((((.((((((....))))))..)))).))).)))))..))))))..(((..((.(.....).))..)))...((((.....))))....... ( -32.10) >DroSec_CAF1 211335 114 + 1 GAAAUGCAAGUCAGUAAAGUCCUGGAAAAUCCACAAGGAUUUGCUCGGUGAUAGCUGAAAGCUUUCUCCCAUUCCCCACAUUGAGGUUUGGAUUUCCCUUUCGGGGAUUUUCCA ((((.((...(((((...(((((((.((((((....))))))..)))).))).)))))..))))))..(((..((.(.....).))..)))...((((.....))))....... ( -32.40) >DroSim_CAF1 220393 114 + 1 GAAAUGCAAGUCAGUAAAGUCCUGGAAAAUCCACAAGGAUUUGCUCGGUGAUAGCUGAAAGCUUUUUCCCAUUCCCCACAUAGAGGUUUGGAUUUCCCUUUCGGGGAUUUUCCA ((((.((...(((((...(((((((.((((((....))))))..)))).))).)))))..))..))))(((..((.(.....).))..)))...((((.....))))....... ( -31.40) >DroEre_CAF1 211792 114 + 1 GAAAUGCAAGUCGGUAAAGUCGUGGAAAAUCCACAAGGAUUUGCUCGGUGAUAGCUGAAAGCUUUUUCCCACUCCCUACAUCGAGAUUUGGAUUUCCCUUUCGGGGAUUUUCCA .....(((((((((.....))((((.....))))...)))))))..((.((.(((.....)))...)))).(((........)))...((((..((((.....))))...)))) ( -31.40) >DroYak_CAF1 221745 114 + 1 GAAAUGCAAGUCGGUAAAGUCCUGGAAAAUCCACAAGGAUUUGCUCGGUGAUAGCUGAAAGCUUUUUCCCAUUCCCCACAUAGAGGUUUGGAUUUCCCUUUCGGGGAUUUUCCA ((((.((...(((((...(((((((.((((((....))))))..)))).))).)))))..))..))))(((..((.(.....).))..)))...((((.....))))....... ( -30.70) >DroAna_CAF1 192640 106 + 1 GAAAUGCAAGUCAGUAAAUUCCUGGAAAAUCCUCAAGGAUUUGCCUGGCGAUAGAUGAAAGCUUUCCC-------CUCCGCCGAGGG-CGGAUUUCCCUUUCGGGGAUUUUCCA ....(((......))).......(((((((((((((((((((((((((((..((..(((....)))..-------)).))))..)))-)))))...))))..))))))))))). ( -34.90) >consensus GAAAUGCAAGUCAGUAAAGUCCUGGAAAAUCCACAAGGAUUUGCUCGGUGAUAGCUGAAAGCUUUCUCCCAUUCCCCACAUCGAGGUUUGGAUUUCCCUUUCGGGGAUUUUCCA ((((.((...(((((...(((((((.((((((....))))))..)))).))).)))))..)))))).......................(((..((((.....))))...))). (-27.88 = -27.63 + -0.25)


| Location | 18,367,572 – 18,367,678 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 92.14 |
| Mean single sequence MFE | -27.65 |
| Consensus MFE | -23.27 |
| Energy contribution | -23.42 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908025 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18367572 106 - 22407834 G-----AAACAAACAAUGUGCUAGUAUAUUGUUUGCAUAUUUCAUGACCUUUGACAACAAUCAACUACGAGUGGAAAAUCCCCGAAAGGGAAAUCCAAACCUCGAUGUGGG (-----((((((((((((((....)))))))))))....))))....((.((((......))))...((((((((.....(((....)))...))))...))))....)). ( -28.10) >DroSec_CAF1 211409 105 - 1 G-----AAACAAACAAUGUGCUGGUAUAU-GUUUGCAUAUUUCAUGACCUUUGACAACAAUCAACUACGAGUGGAAAAUCCCCGAAAGGGAAAUCCAAACCUCAAUGUGGG .-----...((((((.((((....)))))-)))))((((((.........((((......))))....(((((((.....(((....)))...))))...))))))))).. ( -22.10) >DroSim_CAF1 220467 106 - 1 G-----AAACAAACAAUGUGCUGGUAUAUUGUUUGCAUAUUUCAUGACCUUUGACAACAAUCAACUACGAGUGGAAAAUCCCCGAAAGGGAAAUCCAAACCUCUAUGUGGG (-----((((((((((((((....)))))))))))....)))).......((((......))))(((((((((((.....(((....)))...))))...)))...)))). ( -26.30) >DroEre_CAF1 211866 106 - 1 G-----AAACAAACAAUGUGCUGGUAUAUUGUUUGCAUAUUUCAUGACCUUUGACAACAAUCAACUACGAGUGGAAAAUCCCCGAAAGGGAAAUCCAAAUCUCGAUGUAGG (-----((((((((((((((....)))))))))))....))))....(((((((......))))...((((((((.....(((....)))...))))...))))....))) ( -27.50) >DroYak_CAF1 221819 106 - 1 G-----AAACAAACAAUGUGCUGGUAUAUUGUUUGCAUAUUUCAUGACCUUUGACAACAAUCAACUACGAGUGGAAAAUCCCCGAAAGGGAAAUCCAAACCUCUAUGUGGG (-----((((((((((((((....)))))))))))....)))).......((((......))))(((((((((((.....(((....)))...))))...)))...)))). ( -26.30) >DroAna_CAF1 192708 109 - 1 AAUGGCAAACAAACAAUGUGCCAGCGUAUUGUUUGCAUAUUUCAUGACCUUUGACAACAAUCAACCAGGAGUGGAAAAUCCCCGAAAGGGAAAUCCG-CCCUCGGCGGAG- ....((((((((...((((....)))).))))))))...((((((..(((((((......))))..))).))))))....(((....)))...((((-((...)))))).- ( -35.60) >consensus G_____AAACAAACAAUGUGCUGGUAUAUUGUUUGCAUAUUUCAUGACCUUUGACAACAAUCAACUACGAGUGGAAAAUCCCCGAAAGGGAAAUCCAAACCUCGAUGUGGG .........(((((((((((....)))))))))))...............((((......))))(((((((((((.....(((....)))...))))...)))...)))). (-23.27 = -23.42 + 0.14)


| Location | 18,367,612 – 18,367,714 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -22.76 |
| Consensus MFE | -21.74 |
| Energy contribution | -22.10 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.926558 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18367612 102 + 22407834 CUCGUAGUUGAUUGUUGUCAAAGGUCAUGAAAUAUGCAAACAAUAUACUAGCACAUUGUUUGUUU-----CCGAUCCGAUACGAUUAGCUAAUCGAAGCGAUAAACU- .((((((((((((((.(((...((((..(((....(((((((((..........)))))))))))-----).)))).))))))))))))))..)))...........- ( -23.30) >DroSec_CAF1 211449 101 + 1 CUCGUAGUUGAUUGUUGUCAAAGGUCAUGAAAUAUGCAAAC-AUAUACCAGCACAUUGUUUGUUU-----CCGAUCCGAUACGAUUAGCUAAUCGAAGCGAUAAACU- .((((((((((((((.(((...((((..((((((.((((..-.............)))).)))))-----).)))).))))))))))))))..)))...........- ( -21.86) >DroSim_CAF1 220507 102 + 1 CUCGUAGUUGAUUGUUGUCAAAGGUCAUGAAAUAUGCAAACAAUAUACCAGCACAUUGUUUGUUU-----CCGAUCCGAUACGAUUAGCUAAUCGAAGCGAUAAACU- .((((((((((((((.(((...((((..(((....(((((((((..........)))))))))))-----).)))).))))))))))))))..)))...........- ( -23.30) >DroEre_CAF1 211906 103 + 1 CUCGUAGUUGAUUGUUGUCAAAGGUCAUGAAAUAUGCAAACAAUAUACCAGCACAUUGUUUGUUU-----CCGAUCCGAUACGAUUAGCUAAUCGAAGCGAUAAACUC .((((((((((((((.(((...((((..(((....(((((((((..........)))))))))))-----).)))).))))))))))))))..)))............ ( -23.30) >DroYak_CAF1 221859 102 + 1 CUCGUAGUUGAUUGUUGUCAAAGGUCAUGAAAUAUGCAAACAAUAUACCAGCACAUUGUUUGUUU-----CCGAUCCGAUACGAUUAGCUAAUCGAAGCGAUAAACU- .((((((((((((((.(((...((((..(((....(((((((((..........)))))))))))-----).)))).))))))))))))))..)))...........- ( -23.30) >DroAna_CAF1 192746 107 + 1 CUCCUGGUUGAUUGUUGUCAAAGGUCAUGAAAUAUGCAAACAAUACGCUGGCACAUUGUUUGUUUGCCAUUCGAUCCGAUCCGAUUAGCUAAUCGAAGCGAUAAACU- ....((((((((((..(((...((((..(((....((((((((.(((.((...)).)))))))))))..))))))).))).))))))))))((((...)))).....- ( -21.50) >consensus CUCGUAGUUGAUUGUUGUCAAAGGUCAUGAAAUAUGCAAACAAUAUACCAGCACAUUGUUUGUUU_____CCGAUCCGAUACGAUUAGCUAAUCGAAGCGAUAAACU_ .((((((((((((((.(((...((((.........(((((((((..........))))))))).........)))).))))))))))))))..)))............ (-21.74 = -22.10 + 0.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:13:23 2006