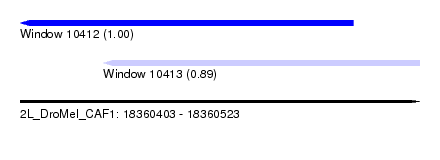
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,360,403 – 18,360,523 |
| Length | 120 |
| Max. P | 0.999239 |

| Location | 18,360,403 – 18,360,503 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 90.24 |
| Mean single sequence MFE | -28.46 |
| Consensus MFE | -23.26 |
| Energy contribution | -24.02 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.50 |
| Structure conservation index | 0.82 |
| SVM decision value | 3.45 |
| SVM RNA-class probability | 0.999239 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
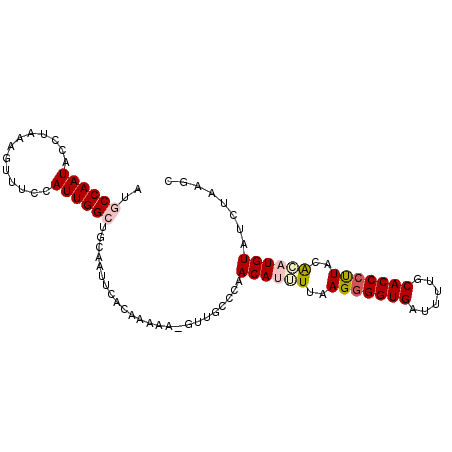
>2L_DroMel_CAF1 18360403 100 - 22407834 CAUUGGCUGCAAUUCACAAAAAGGUUUCCCAACAUUUUAAGGGGUGAUUUUGCACCCUUACACAUGUAUCUAAGCCAAAUGUUGCAGCGACAACAAUGUU (((((((((((((.........(((((....((((....(((((((......)))))))....))))....)))))....)))))))).....))))).. ( -27.72) >DroSec_CAF1 204249 99 - 1 CAUUGGCUGCAAUUCACAAAAA-GUUGCCCAACAUGUUAAGGGGUGAUUUUGCACCCUUACACAUGUAUCUAAGCCAAAUGUUGCAGCGACAACAAUGUU ..((((((((((((.......)-)))))...((((((..(((((((......)))))))..)))))).....)))))).(((((......)))))..... ( -31.70) >DroSim_CAF1 212757 99 - 1 CAUUGGCUGCAAUUCACAAAAA-GUUUCCCAACAUGUUAAGGGGUGAUUUUGCACCCUUACACAUGUAUCUAAACCAAAUGUUGCAGCGACAACAAUGUU (((((((((((((.........-((((....((((((..(((((((......)))))))..))))))....)))).....)))))))).....))))).. ( -29.44) >DroEre_CAF1 204651 95 - 1 CAUUGGCUGAAAUUCCAAAAAA-GUUGCCCAACAAUUUUAG-GGUGAAUUUGCACCCUUACAUAUGUAUCUAAGCCAAAUGUUGCAGCGAC---AAUGUU ..((((........))))....-(((((.(((((.....((-((((......))))))(((....)))...........)))))..)))))---...... ( -21.80) >DroYak_CAF1 214563 99 - 1 CAUUGGCUGCAAUUCACAAAAA-GUUGCUAAACAAUUUUAGGGGUGAAUUUGCACCCCUGCGAAUGUAUCUAAGCAAAAUGUUGCAGCGACAACAAUGUU (((((((((((((.........-.(((((..(((.((.((((((((......)))))))).)).))).....)))))...)))))))).....))))).. ( -31.62) >consensus CAUUGGCUGCAAUUCACAAAAA_GUUGCCCAACAUUUUAAGGGGUGAUUUUGCACCCUUACACAUGUAUCUAAGCCAAAUGUUGCAGCGACAACAAUGUU (((((((((((((..........(((.....((((((..(((((((......)))))))..)))))).....))).....)))))))).....))))).. (-23.26 = -24.02 + 0.76)


| Location | 18,360,428 – 18,360,523 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 89.51 |
| Mean single sequence MFE | -20.37 |
| Consensus MFE | -16.08 |
| Energy contribution | -17.00 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.891249 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18360428 95 - 22407834 AUGCCAAUACCUACAGUUUCCAUUGGCUGCAAUUCACAAAAAGGUUUCCCAACAUUUUAAGGGGUGAUUUUGCACCCUUACACAUGUAUCUAAGC ..((((((.............)))))).((............((....)).((((....(((((((......)))))))....))))......)) ( -18.52) >DroSec_CAF1 204274 94 - 1 AUGCCAAUACCUAGAGUUUCCAUUGGCUGCAAUUCACAAAAA-GUUGCCCAACAUGUUAAGGGGUGAUUUUGCACCCUUACACAUGUAUCUAAGC ..((((((.............)))))).((((((.......)-)))))...((((((..(((((((......)))))))..))))))........ ( -25.32) >DroSim_CAF1 212782 94 - 1 AUGCCAAUACCUAGAGUUUCCAUUGGCUGCAAUUCACAAAAA-GUUUCCCAACAUGUUAAGGGGUGAUUUUGCACCCUUACACAUGUAUCUAAAC ..((((((.............))))))...............-........((((((..(((((((......)))))))..))))))........ ( -21.82) >DroEre_CAF1 204673 93 - 1 AUCCCAAUACCUAAAGUUUCCAUUGGCUGAAAUUCCAAAAAA-GUUGCCCAACAAUUUUAG-GGUGAAUUUGCACCCUUACAUAUGUAUCUAAGC ......((((.((.........((((........)))).(((-((((.....)))))))((-((((......))))))....)).))))...... ( -13.60) >DroYak_CAF1 214588 94 - 1 AUCCCAAUAUCUAAAGUUUCCAUUGGCUGCAAUUCACAAAAA-GUUGCUAAACAAUUUUAGGGGUGAAUUUGCACCCCUGCGAAUGUAUCUAAGC ......................((((.((((.(((.(..(((-((((.....)))))))(((((((......)))))))).))))))).)))).. ( -22.60) >consensus AUGCCAAUACCUAAAGUUUCCAUUGGCUGCAAUUCACAAAAA_GUUGCCCAACAUUUUAAGGGGUGAUUUUGCACCCUUACACAUGUAUCUAAGC ..((((((.............))))))........................((((((..(((((((......)))))))..))))))........ (-16.08 = -17.00 + 0.92)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:13:19 2006