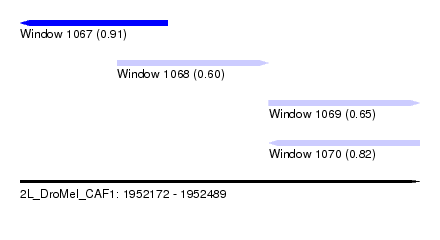
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,952,172 – 1,952,489 |
| Length | 317 |
| Max. P | 0.906725 |

| Location | 1,952,172 – 1,952,289 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.22 |
| Mean single sequence MFE | -55.95 |
| Consensus MFE | -40.10 |
| Energy contribution | -42.35 |
| Covariance contribution | 2.25 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.906725 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1952172 117 - 22407834 CUUCACCGCAUCCCUGAAUGCCAGCUUGCAUCACUCGCAGUCGCUGAGAAGAAAUCUGGGUCAUCCACUGGCCGCGGCAGCUGCCGUGGCGGCAGUGGCCCAAUCCCAGGCGG---UGCC ...((((((............((((((((.......))))..)))).........(((((....((((((((((((((....))))))))..))))))......)))))))))---)).. ( -52.70) >DroEre_CAF1 3821 117 - 1 CUUCACCGCCUCCCUGAACGCCAGCUUGCAUCACUCGCAGUCGCUGAGAAGGAAUCUGGGCCAUCCUCUGGCCGCGGCAGCUGCGGUGGCGGCGGCGGCCCAGUCCCAGGCGG---UGCC ...((((((((..(((..((((.(((.((..(((.((((((.(((((((.....))).(((((.....))))).)))).))))))))))))))))))...)))....))))))---)).. ( -57.10) >DroYak_CAF1 1393 117 - 1 CUUCACCGCCUCCCUGAACGCCAGCCUGCAUCACUCGCAGUCGCUGCGAAGGAAUCUGGGUCAUCCGCUGGCCGCGGCUGCUGCCGUGGCGGCGGCUGCCCAAUCCCAGGCGG---UGCC ...((((((((.(((...(((.(((((((.......))))..)))))).)))....((((.((.((((((.(((((((....))))))))))))).)))))).....))))))---)).. ( -63.70) >DroAna_CAF1 24008 120 - 1 CUUCACCGCCUCGCUGAACGCCAGUCUCCACCACUCGCAGUCGCUGCGCCGCAAUCUGGGCCAUCCUUUGGCUGCAGCAGCCGCCGCAGCGGCCGUGGCACAGUCCCAGGCGGUAGUGCC ....(((((((.((((...((((((.......))).((.((((((((((.((...((((((((.....))))).)))..)).).))))))))).))))).))))...)))))))...... ( -50.30) >consensus CUUCACCGCCUCCCUGAACGCCAGCCUGCAUCACUCGCAGUCGCUGAGAAGGAAUCUGGGCCAUCCUCUGGCCGCGGCAGCUGCCGUGGCGGCGGCGGCCCAAUCCCAGGCGG___UGCC .....((((((..........((((((((.......))))..))))....((...((((((((((...(.((((((((....)))))))).).))))))))))..))))))))....... (-40.10 = -42.35 + 2.25)

| Location | 1,952,249 – 1,952,369 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.92 |
| Mean single sequence MFE | -55.80 |
| Consensus MFE | -45.59 |
| Energy contribution | -47.40 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.599297 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1952249 120 + 22407834 CUGCGAGUGAUGCAAGCUGGCAUUCAGGGAUGCGGUGAAGCUGCUCAGGUGGGCGGCGCUGGCCAGCGAUUGCUGUUGGGCGGUAAGGGCUGCAUAUUGGGCGGCAGCAGCGGCGCACAU ......(((.(((..(((((((((....)))))......(((((((((....(((((((((.((((((.....)))))).))))....)))))...)))))))))..)))).)))))).. ( -54.60) >DroEre_CAF1 3898 120 + 1 CUGCGAGUGAUGCAAGCUGGCGUUCAGGGAGGCGGUGAAGCUGCUCAGGUGGGCGGCGCUGGCCAGCGAUUGCUGUUGGGCGGUCAGGGCCGCAUACUGGGCGGCAGCGGCGGCGCACAU ......(((.(((..((((.(((((((....(((((...(((((((....)))))))((((.((((((.....)))))).))))....)))))...))))))).))))....)))))).. ( -61.60) >DroYak_CAF1 1470 120 + 1 CUGCGAGUGAUGCAGGCUGGCGUUCAGGGAGGCGGUGAAGCUGCUCAGGUGGGCGGCGCUGGCCAGCGAUUGCUGUUGGGCGGUCAGGGCCGCGUAUUGGGCGGCGGCGGCGGCGCACAU .((((.....(((..((((.(((((((..(.(((((...(((((((....)))))))((((.((((((.....)))))).))))....))))).).))))))).)))).))).))))... ( -59.60) >DroAna_CAF1 24088 111 + 1 CUGCGAGUGGUGGAGACUGGCGUUCAGCGAGGCGGUGAAGCUGCUCAGGUGGG---------UCAGCGACUGCUGCUGGGCAGUCAGGGCGGCGUACUGGGCGGCGGCGGCGGCACACAU ......(((((.(...((((((((((((..((((((...(((((((....)))---------.)))).))))))))))))).)))))(.((.(((.....))).)).)..).)).))).. ( -47.40) >consensus CUGCGAGUGAUGCAAGCUGGCGUUCAGGGAGGCGGUGAAGCUGCUCAGGUGGGCGGCGCUGGCCAGCGAUUGCUGUUGGGCGGUCAGGGCCGCAUACUGGGCGGCAGCGGCGGCGCACAU ......(((.(((..((((.(((((((..(.(((((...(((((((....)))))))((((.((((((.....)))))).))))....))))).).))))))).)))).))).))).... (-45.59 = -47.40 + 1.81)

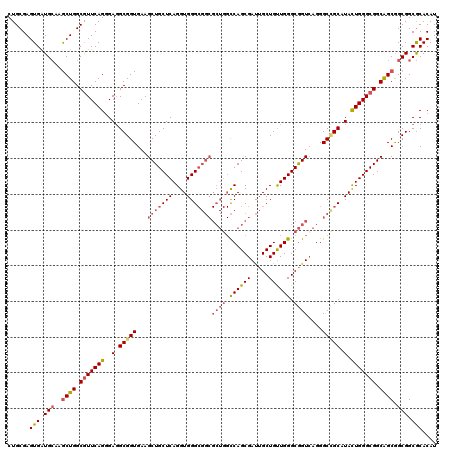
| Location | 1,952,369 – 1,952,489 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.33 |
| Mean single sequence MFE | -51.55 |
| Consensus MFE | -43.20 |
| Energy contribution | -43.32 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.649746 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1952369 120 + 22407834 CAGCUGAUUGGGUGCAUAGUACAGCAGGGGAUACUGCGAGAGCAGCUCCUGGUGGCGGGAGCUCACAAACUGCUGCACAGCAGCGGUGCUCGGCGGCGAGGCCACCGAUGAAGUGGCGCC ..((((.....(((((((((....((((((...((((....))))))))))(((((....)).)))..)))).)))))..))))(((((((((.(((...))).)))).......))))) ( -54.51) >DroEre_CAF1 4018 120 + 1 CAGCUGAUUGGGUGCAUAGUAGAGCAGGGGAUACUGCGAGAGCAGCUCCUGGUGGCGGGAGCUCACGAACUGCUGCACAGCGGCGGUGCUCGGAGGCGAGGCCACCGACGUGGUGGCGCC ..((((.....(((((..((((..((((((...((((....))))))))))(((((....)).)))...)))))))))..))))(((.((((....))))((((((.....))))))))) ( -56.70) >DroYak_CAF1 1590 120 + 1 CAGCUGAUUGGGUGCAUAGUACAGCAGGGGAUACUGCGAGAGCAGCUCCUGGUGGCGGGAGCUCACGAACUGCUGCACUGCAGCGGUGCUCGGCGGCGAGGCCACCGAUGCAGUGGCGCC ..........(((((......((((((((((..((((....)))).)))).(((((....)).)))...))))))(((((((.(((((((((....))))..))))).)))))))))))) ( -60.20) >DroAna_CAF1 24199 102 + 1 CAGCUGGUUGGGAGCGUAGUACAGCAGGGGAUACUGGGAAAGCAGCUCCUGGUGGCGGGUGUUUACAAAUUGCUGCACGGCUGUGG------------------CCGAGUUGGUGGCGCC ((((...(((..((((..((.((.((((((...(((......))))))))).))))...))))..)))...))))...(((.((.(------------------((.....))).))))) ( -34.80) >consensus CAGCUGAUUGGGUGCAUAGUACAGCAGGGGAUACUGCGAGAGCAGCUCCUGGUGGCGGGAGCUCACAAACUGCUGCACAGCAGCGGUGCUCGGCGGCGAGGCCACCGAUGUAGUGGCGCC ..........(((((......((((((((((..((((....)))).)))).(((((....)).)))...))))))(((.(((.(((((((((....))))..))))).))).)))))))) (-43.20 = -43.32 + 0.12)

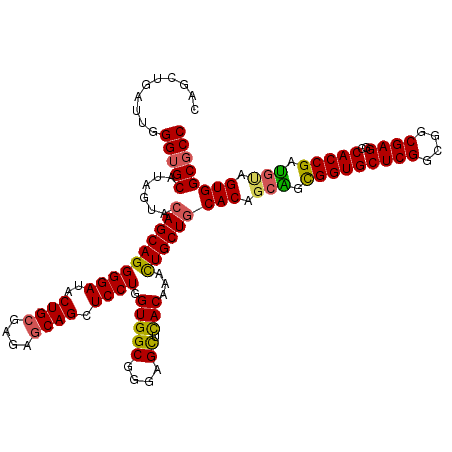
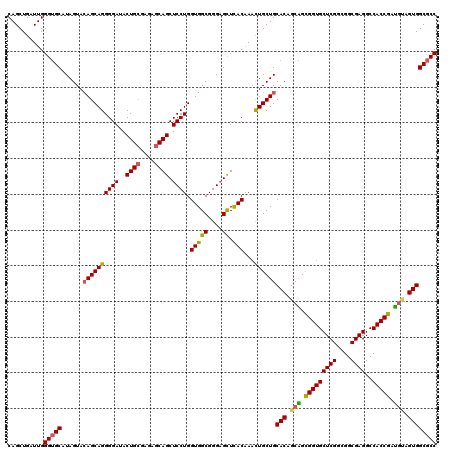
| Location | 1,952,369 – 1,952,489 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.33 |
| Mean single sequence MFE | -43.60 |
| Consensus MFE | -32.97 |
| Energy contribution | -34.98 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.816500 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1952369 120 - 22407834 GGCGCCACUUCAUCGGUGGCCUCGCCGCCGAGCACCGCUGCUGUGCAGCAGUUUGUGAGCUCCCGCCACCAGGAGCUGCUCUCGCAGUAUCCCCUGCUGUACUAUGCACCCAAUCAGCUG (((((.......((((((((...)))))))).....))(((.(((((((((...(((.((....)))))..((((((((....))))).))).)))))))))...)))........))). ( -48.90) >DroEre_CAF1 4018 120 - 1 GGCGCCACCACGUCGGUGGCCUCGCCUCCGAGCACCGCCGCUGUGCAGCAGUUCGUGAGCUCCCGCCACCAGGAGCUGCUCUCGCAGUAUCCCCUGCUCUACUAUGCACCCAAUCAGCUG (((((((((.....))))))((((....))))....)))((((((((((((((((((.((....)))))...)))))))....((((......)))).......)))))......))).. ( -46.70) >DroYak_CAF1 1590 120 - 1 GGCGCCACUGCAUCGGUGGCCUCGCCGCCGAGCACCGCUGCAGUGCAGCAGUUCGUGAGCUCCCGCCACCAGGAGCUGCUCUCGCAGUAUCCCCUGCUGUACUAUGCACCCAAUCAGCUG (((((...(((.((((((((...)))))))))))..))(((((((((((((...(((.((....)))))..((((((((....))))).))).)))))))))..))))........))). ( -53.40) >DroAna_CAF1 24199 102 - 1 GGCGCCACCAACUCGG------------------CCACAGCCGUGCAGCAAUUUGUAAACACCCGCCACCAGGAGCUGCUUUCCCAGUAUCCCCUGCUGUACUACGCUCCCAACCAGCUG ((((((........))------------------)....)))((((((((...((....))..........(((((((......)))).)))..))))))))...(((.......))).. ( -25.40) >consensus GGCGCCACCACAUCGGUGGCCUCGCCGCCGAGCACCGCUGCUGUGCAGCAGUUCGUGAGCUCCCGCCACCAGGAGCUGCUCUCGCAGUAUCCCCUGCUGUACUAUGCACCCAAUCAGCUG (((((((((.....))))))...................((.(((((((((...(((.((....)))))..((((((((....))))).))).)))))))))...)).........))). (-32.97 = -34.98 + 2.00)

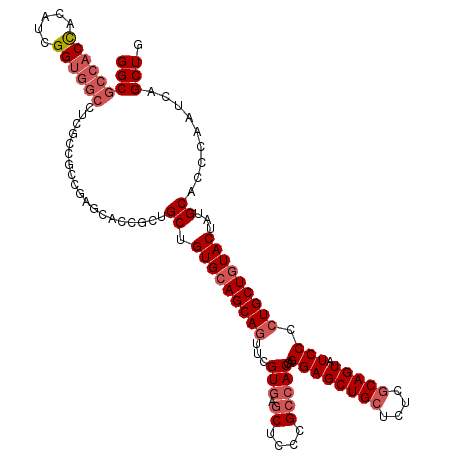
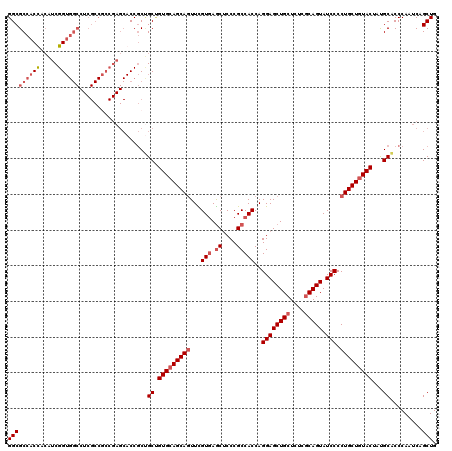
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:42:03 2006