
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,336,287 – 18,336,385 |
| Length | 98 |
| Max. P | 0.613148 |

| Location | 18,336,287 – 18,336,385 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 80.05 |
| Mean single sequence MFE | -25.96 |
| Consensus MFE | -15.84 |
| Energy contribution | -16.51 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.613148 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
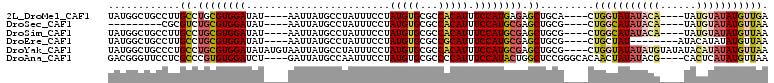
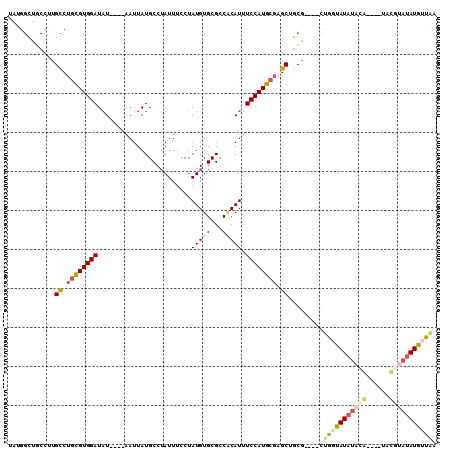
>2L_DroMel_CAF1 18336287 98 + 22407834 UAUGGCUGCCUUGCCUGCGUGGAUAU----AAUUAUGCCUAUUUCCUAUGUGCGCCACAUUUCCAUGAGAGCUGCA----CUGGUAUAUACA----UAUGUAUAUGUUGA ...(((......))).(((((.((((----(..((((((..........(((((((.(((....))).).)).)))----).))))))....----))))).)))))... ( -21.50) >DroSec_CAF1 179991 89 + 1 ---------CGCGUCUGCGUGGAUAU----AAUUAUGCCUAUUUCCUAUGUGCGCCACAUUUCCAUGCGAGCUGCG----CUGGCAUAUACA----UAUGUAUAUGUUAA ---------((((..((((((((...----.....(((.(((.....))).))).......))))))))..).)))----.((((((((((.----...)))))))))). ( -27.96) >DroSim_CAF1 188171 98 + 1 UAUGGCUGCCUUGCCUGCGUGGAUAU----AAUUAUGCCUAUUUCCUAUGUGCGCCACAUUUCCAUGCGAGCUGCG----CUGGCAUAUACA----UAUGUAUAUGUUAA ...(((.((...((.((((((((...----.....(((.(((.....))).))).......)))))))).)).)))----))(((((((((.----...))))))))).. ( -29.16) >DroEre_CAF1 181331 94 + 1 UAUGGCUGCCUUGCCUGCGUGGAUAU----AAUUAUGCCUAUUUCCUAUGUGCGCCGCAUUUCCAUGCGAGCUGCG----CUGCUAU--------AUACAUAUAUGUUAA ((((((.((...((.((((((((...----.....(((.(((.....))).))).......)))))))).))...)----).)))))--------).............. ( -25.26) >DroYak_CAF1 191129 106 + 1 UAUGGCUGCCCUGCCUGCGUGGAUAUAUGUAAUUAUGCCUAUUUCCUAUGUGCGCCACAUUUCCAUGCGAGCUGCG----CUGGUAUAUAUGUAUAUACAUAUAUGUUAA ...(((.((...((.((((((((...((((.....(((.(((.....))).)))..)))).)))))))).)).)))----))..(((((((((....))))))))).... ( -28.60) >DroAna_CAF1 163681 102 + 1 GACGGGUUCCUCGCCCGUGUGGAUCU----GAUUAUGCCAAUUUCCUAUGUGCGCCCCAUUUCCAUACUGGCUCCGGGCACAACUAUAUACG----CACUCAUAUGUUAA (((((((.....))))(((((.....----......((((........)).))((((.....((.....))....))))...........))----)))......))).. ( -23.30) >consensus UAUGGCUGCCUUGCCUGCGUGGAUAU____AAUUAUGCCUAUUUCCUAUGUGCGCCACAUUUCCAUGCGAGCUGCG____CUGGUAUAUACA____UACGUAUAUGUUAA ............((.((((((((........................(((((...))))).)))))))).)).........(((((((((((......))))))))))). (-15.84 = -16.51 + 0.67)

| Location | 18,336,287 – 18,336,385 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 80.05 |
| Mean single sequence MFE | -26.25 |
| Consensus MFE | -13.77 |
| Energy contribution | -15.13 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.52 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.515670 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
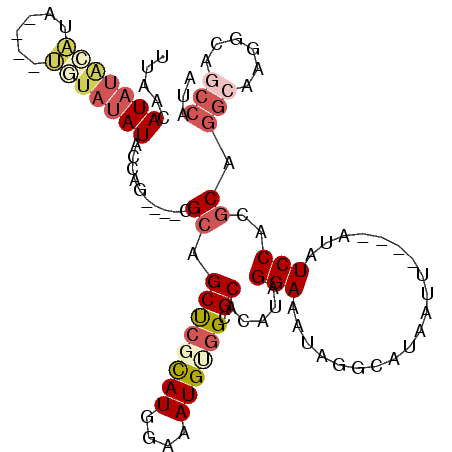
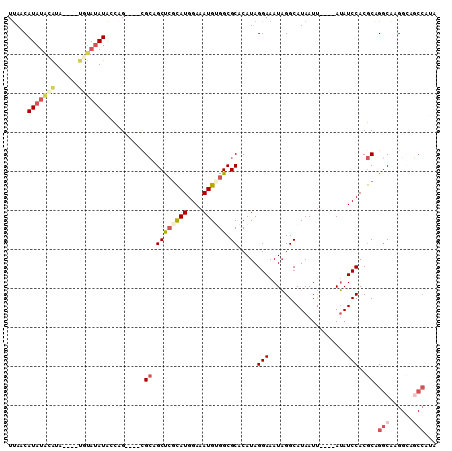
>2L_DroMel_CAF1 18336287 98 - 22407834 UCAACAUAUACAUA----UGUAUAUACCAG----UGCAGCUCUCAUGGAAAUGUGGCGCACAUAGGAAAUAGGCAUAAUU----AUAUCCACGCAGGCAAGGCAGCCAUA .....((((((...----.))))))....(----(((.(((..(((....))).)))))))...(((.((((......))----)).))).....(((......)))... ( -24.90) >DroSec_CAF1 179991 89 - 1 UUAACAUAUACAUA----UGUAUAUGCCAG----CGCAGCUCGCAUGGAAAUGUGGCGCACAUAGGAAAUAGGCAUAAUU----AUAUCCACGCAGACGCG--------- ....(((((((...----.)))))))...(----(((.((((((((....)))))).)).....(((.((((......))----)).))).....).))).--------- ( -24.70) >DroSim_CAF1 188171 98 - 1 UUAACAUAUACAUA----UGUAUAUGCCAG----CGCAGCUCGCAUGGAAAUGUGGCGCACAUAGGAAAUAGGCAUAAUU----AUAUCCACGCAGGCAAGGCAGCCAUA ....(((((((...----.)))))))...(----(((.((.(((((....))))))))).....(((.((((......))----)).)))..)).(((......)))... ( -29.30) >DroEre_CAF1 181331 94 - 1 UUAACAUAUAUGUAU--------AUAGCAG----CGCAGCUCGCAUGGAAAUGCGGCGCACAUAGGAAAUAGGCAUAAUU----AUAUCCACGCAGGCAAGGCAGCCAUA ...............--------...((.(----(((....(((((....))))))))).....(((.((((......))----)).)))..)).(((......)))... ( -25.60) >DroYak_CAF1 191129 106 - 1 UUAACAUAUAUGUAUAUACAUAUAUACCAG----CGCAGCUCGCAUGGAAAUGUGGCGCACAUAGGAAAUAGGCAUAAUUACAUAUAUCCACGCAGGCAGGGCAGCCAUA .....((((((((....))))))))....(----(((.((.(((((....))))))))).....(((.(((............))).)))..)).(((......)))... ( -27.00) >DroAna_CAF1 163681 102 - 1 UUAACAUAUGAGUG----CGUAUAUAGUUGUGCCCGGAGCCAGUAUGGAAAUGGGGCGCACAUAGGAAAUUGGCAUAAUC----AGAUCCACACGGGCGAGGAACCCGUC ...........(((----((((((....))).((((...((.....))...))))))))))...(((..((((.....))----)).)))..(((((.......))))). ( -26.00) >consensus UUAACAUAUACAUA____UGUAUAUACCAG____CGCAGCUCGCAUGGAAAUGUGGCGCACAUAGGAAAUAGGCAUAAUU____AUAUCCACGCAGGCAAGGCAGCCAUA .....(((((((......)))))))..........((.((((((((....)))))).)).....(((....................)))..)).(((......)))... (-13.77 = -15.13 + 1.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:13:05 2006