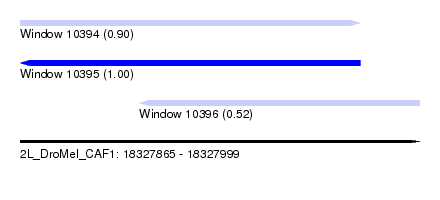
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,327,865 – 18,327,999 |
| Length | 134 |
| Max. P | 0.996776 |

| Location | 18,327,865 – 18,327,979 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 90.98 |
| Mean single sequence MFE | -32.94 |
| Consensus MFE | -23.18 |
| Energy contribution | -24.22 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.21 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.895990 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18327865 114 + 22407834 UUUUUUUGUUGUGCAGCAGCUACUCAAACUAUAGGCGGCACAAACAAAUCUCCGGCUUUGCUAUAGAAAAGCUACUUCCAUGGCAAGUUGCCGCUACUAAAGCCAAAGCCAAAG- ....((((((((((....(((............))).)))).)))))).....(((((((((.(((....((.(((((....).)))).)).....))).)).)))))))....- ( -30.10) >DroSec_CAF1 171631 112 + 1 ---UUUUGUUGUGCAGUAGCUACUCAAACUAUAGCCGGCACAAACAAAUCUCCGGCUUUGCUAUAGAAAAGCUACUUCCAUGGCAAGUUGGCGCUACUAAAGCUAAAGCCAAAGC ---.(((((..((.(((((((..((.......((((((.............))))))........))..)))))))..))..)))))((((((((.....)))....)))))... ( -33.08) >DroSim_CAF1 179814 112 + 1 ---UUUUGUUGUGCAGUAGCUACUCAAACUAUAGCCGGCACAAACAAAUCUCCGGCUUUGCUAUAGAAAAGCUACUUCCAUGGCAAGUUGGCGCUACUAAAGCCAAAGCCAAAGC ---.(((((..((.(((((((..((.......((((((.............))))))........))..)))))))..))..)))))((((((((.....)))....)))))... ( -33.08) >DroEre_CAF1 173119 112 + 1 ---CUUUUUUGUGCAGCAGCUACUCAAACUGUAGCUGGCACAAACAAAUCUCCGGCUUUGCUAUAGAAAAGCUACUUCCAUGGCAAGUUGCCGCUGCUAAAGCCAAAGCCAAAGC ---....(((((((..(((((((.......)))))))))))))).........(((((((((.(((...(((.........(((.....)))))).))).)).)))))))..... ( -38.80) >DroYak_CAF1 183036 103 + 1 ---UUUUUUGGUGCAGCAGCUACUCAAACUAUAGCUGGUACAAACAAAUCUCCGGCUUUGCUAUAGAAAAGCUACUUCCAUGGCAAGUUGCCGCUACUAAAGCCAA--------- ---..(((((((((.((((((...........((((((.............)))))).((((((.(((.......))).)))))))))))).)).)))))))....--------- ( -29.62) >consensus ___UUUUGUUGUGCAGCAGCUACUCAAACUAUAGCCGGCACAAACAAAUCUCCGGCUUUGCUAUAGAAAAGCUACUUCCAUGGCAAGUUGCCGCUACUAAAGCCAAAGCCAAAGC ........((((((....((((.........))))..))))))..........(((((((((.(((...(((.(((((....).))))....))).))).))).))))))..... (-23.18 = -24.22 + 1.04)

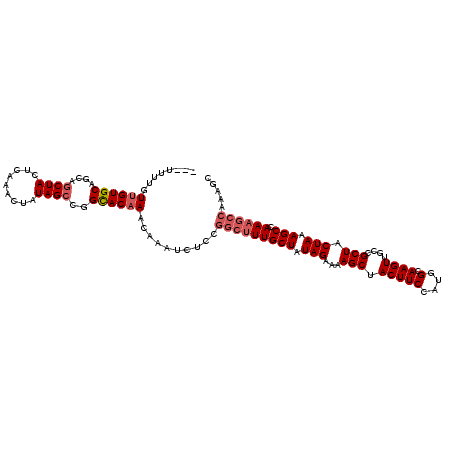
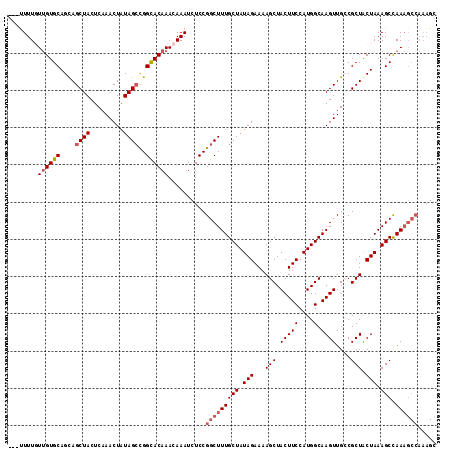
| Location | 18,327,865 – 18,327,979 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 90.98 |
| Mean single sequence MFE | -39.22 |
| Consensus MFE | -30.78 |
| Energy contribution | -33.38 |
| Covariance contribution | 2.60 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.43 |
| Structure conservation index | 0.78 |
| SVM decision value | 2.75 |
| SVM RNA-class probability | 0.996776 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18327865 114 - 22407834 -CUUUGGCUUUGGCUUUAGUAGCGGCAACUUGCCAUGGAAGUAGCUUUUCUAUAGCAAAGCCGGAGAUUUGUUUGUGCCGCCUAUAGUUUGAGUAGCUGCUGCACAACAAAAAAA -((((((((((((((.....)))(((.....)))(((((((.....)))))))..))))))))))).((((((.((((.((((((.......))))..)).)))))))))).... ( -40.20) >DroSec_CAF1 171631 112 - 1 GCUUUGGCUUUAGCUUUAGUAGCGCCAACUUGCCAUGGAAGUAGCUUUUCUAUAGCAAAGCCGGAGAUUUGUUUGUGCCGGCUAUAGUUUGAGUAGCUACUGCACAACAAAA--- .((((((((((.(((.(((.(((.(((........))).....)))...))).))))))))))))).((((((.((((.((((((.......))))))...)))))))))).--- ( -39.80) >DroSim_CAF1 179814 112 - 1 GCUUUGGCUUUGGCUUUAGUAGCGCCAACUUGCCAUGGAAGUAGCUUUUCUAUAGCAAAGCCGGAGAUUUGUUUGUGCCGGCUAUAGUUUGAGUAGCUACUGCACAACAAAA--- .(((((((((((((.........))))).((((.(((((((.....))))))).)))))))))))).((((((.((((.((((((.......))))))...)))))))))).--- ( -40.00) >DroEre_CAF1 173119 112 - 1 GCUUUGGCUUUGGCUUUAGCAGCGGCAACUUGCCAUGGAAGUAGCUUUUCUAUAGCAAAGCCGGAGAUUUGUUUGUGCCAGCUACAGUUUGAGUAGCUGCUGCACAAAAAAG--- .((((((((((((((.....)))(((.....)))(((((((.....)))))))..))))))))))).....((((((((((((((.......)))))))..)))))))....--- ( -44.60) >DroYak_CAF1 183036 103 - 1 ---------UUGGCUUUAGUAGCGGCAACUUGCCAUGGAAGUAGCUUUUCUAUAGCAAAGCCGGAGAUUUGUUUGUACCAGCUAUAGUUUGAGUAGCUGCUGCACCAAAAAA--- ---------((((((((.((...(((.....)))(((((((.....))))))).))))))))))...((((..((((.(((((((.......))))))).)))).))))...--- ( -31.50) >consensus GCUUUGGCUUUGGCUUUAGUAGCGGCAACUUGCCAUGGAAGUAGCUUUUCUAUAGCAAAGCCGGAGAUUUGUUUGUGCCGGCUAUAGUUUGAGUAGCUGCUGCACAACAAAA___ .((((((((((.(((.(((.(((....((((.(....))))).)))...))).))))))))))))).((((((.(((((((((((.......)))))))..)))))))))).... (-30.78 = -33.38 + 2.60)

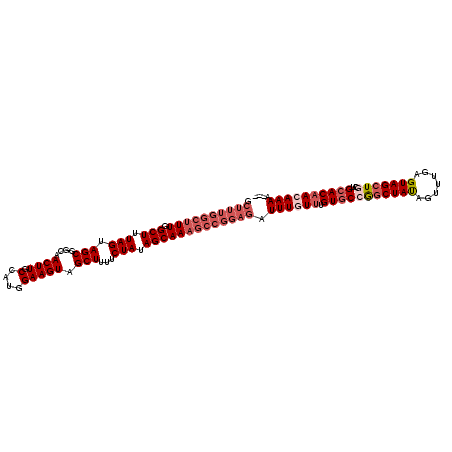
| Location | 18,327,905 – 18,327,999 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 88.52 |
| Mean single sequence MFE | -35.16 |
| Consensus MFE | -22.36 |
| Energy contribution | -25.88 |
| Covariance contribution | 3.52 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.64 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.520069 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18327905 94 - 22407834 CUCUAAAGUGGGUCGAUUUG-------------CUUUGGCUUUGGCUUUAGUAGCGGCAACUUGCCAUGGAAGUAGCUUUUCUAUAGCAAAGCCGGAGAUUUGUUUG ..((((((..((....))..-------------))))))((((((((((.((...(((.....)))(((((((.....))))))).))))))))))))......... ( -29.50) >DroSec_CAF1 171668 107 - 1 CUCCAAAGUGGGUCGAUUUGGCUUUGGCUACGGCUUUGGCUUUAGCUUUAGUAGCGCCAACUUGCCAUGGAAGUAGCUUUUCUAUAGCAAAGCCGGAGAUUUGUUUG .....(((..((((..((((((((((.(((.(((.(((((....(((.....))))))))...))).((((((.....)))))))))))))))))))))))..))). ( -35.80) >DroSim_CAF1 179851 107 - 1 CUCCAAAGUGGGUCGAUUUGGCUUUGGCUACGGCUUUGGCUUUGGCUUUAGUAGCGCCAACUUGCCAUGGAAGUAGCUUUUCUAUAGCAAAGCCGGAGAUUUGUUUG .....(((..((((..(((((((((((((((..((.((((.(((((.........)))))...)))).))..)))))..........))))))))))))))..))). ( -37.20) >DroEre_CAF1 173156 106 - 1 CGCCAAAGUGGGUCGAUUUG-CUUUGGCUUUGGCUUUGGCUUUGGCUUUAGCAGCGGCAACUUGCCAUGGAAGUAGCUUUUCUAUAGCAAAGCCGGAGAUUUGUUUG .(((((((((((((((....-..)))))))..))))))))(((((((((.((...(((.....)))(((((((.....))))))).))))))))))).......... ( -40.10) >DroYak_CAF1 183073 94 - 1 CGCCAAAGUGGGUCGAUUUG-CUUUGGCU------------UUGGCUUUAGUAGCGGCAACUUGCCAUGGAAGUAGCUUUUCUAUAGCAAAGCCGGAGAUUUGUUUG .(((((((..((....))..-)))))))(------------((((((((.((...(((.....)))(((((((.....))))))).))))))))))).......... ( -33.20) >consensus CUCCAAAGUGGGUCGAUUUG_CUUUGGCU__GGCUUUGGCUUUGGCUUUAGUAGCGGCAACUUGCCAUGGAAGUAGCUUUUCUAUAGCAAAGCCGGAGAUUUGUUUG ..(((((((.((((((.......))))))...)))))))((((((((((.((...(((.....)))(((((((.....))))))).))))))))))))......... (-22.36 = -25.88 + 3.52)

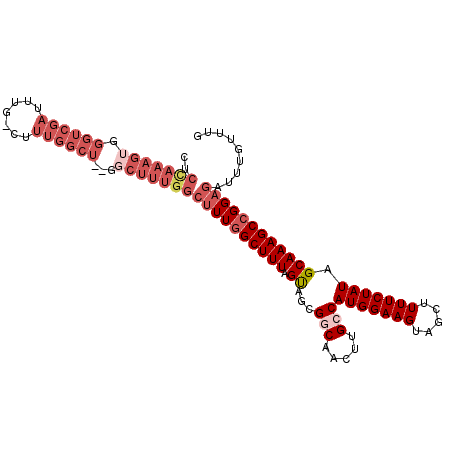
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:13:03 2006